+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2018 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM reconstruction of endophilin N-BAR tubes | |||||||||
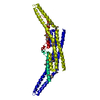
 Map data Map data | This is a map of an endophilin-lipid tube. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | endocytosis / BAR / N-BAR / membrane remodeling | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 21.0 Å | |||||||||
 Authors Authors | Mizuno N / Jao CC / Langen R / Steven AC | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2010 Journal: J Biol Chem / Year: 2010Title: Multiple modes of endophilin-mediated conversion of lipid vesicles into coated tubes: implications for synaptic endocytosis. Authors: Naoko Mizuno / Christine C Jao / Ralf Langen / Alasdair C Steven /  Abstract: Endophilin A1 is a BAR (Bin/amphiphysin/Rvs) protein abundant in neural synapses that senses and induces membrane curvature, contributing to neck formation in presynaptic endocytic vesicles. To ...Endophilin A1 is a BAR (Bin/amphiphysin/Rvs) protein abundant in neural synapses that senses and induces membrane curvature, contributing to neck formation in presynaptic endocytic vesicles. To investigate its role in membrane remodeling, we used cryoelectron microscopy to characterize structural changes induced in lipid vesicles by exposure to endophilin. The vesicles convert rapidly to coated tubules whose morphology reflects the local concentration of endophilin. Their diameters and curvature resemble those of synaptic vesicles in situ. Three-dimensional reconstructions of quasicylindrical tubes revealed arrays of BAR dimers, flanked by densities that we equate with amphipathic helices whose folding and membrane insertion were attested by EPR. We also observed the compression of bulbous coated tubes into 70-A-wide cylindrical micelles, which appear to mimic the penultimate (hemi-fission) stage of endocytosis. Our findings suggest that the adaptability of endophilin-lipid interactions underlies dynamic changes of endocytic membranes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2018.map.gz emd_2018.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2018-v30.xml emd-2018-v30.xml emd-2018.xml emd-2018.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD2018.png EMD2018.png | 99.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2018 http://ftp.pdbj.org/pub/emdb/structures/EMD-2018 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2018 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2018 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2018.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2018.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of an endophilin-lipid tube. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Endophilin N-BAR domain-lipid tube
| Entire | Name: Endophilin N-BAR domain-lipid tube |
|---|---|
| Components |
|
-Supramolecule #1000: Endophilin N-BAR domain-lipid tube
| Supramolecule | Name: Endophilin N-BAR domain-lipid tube / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Macromolecule #1: endophilin BAR domain
| Macromolecule | Name: endophilin BAR domain / type: protein_or_peptide / ID: 1 / Name.synonym: endophilin / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 20 mM HEPES, 100 mM NaCl |
|---|---|
| Grid | Details: Quantifoil 300 mesh grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: OTHER / Details: Vitrification instrument: Vitrobot |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at x 100,000 magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 66000 |
| Sample stage | Specimen holder: Side-entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 21.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER, IHRSR |
|---|---|
| CTF correction | Details: Phase flipping |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)