+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of Acs1 filament determined by FilamentID | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | metabolic enzyme / filament / cryoEM / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationacid-ammonia (or amide) ligase activity / acetate-CoA ligase / acetate-CoA ligase activity / acetyl-CoA biosynthetic process from acetate / AMP binding / ATP binding / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Hugener J / Xu J / Wettstein R / Ioannidi L / Velikov D / Wollweber F / Henggeler A / Matos J / Pilhofer M | |||||||||
| Funding support |  Switzerland, European Union, 2 items Switzerland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis. Authors: Jannik Hugener / Jingwei Xu / Rahel Wettstein / Lydia Ioannidi / Daniel Velikov / Florian Wollweber / Adrian Henggeler / Joao Matos / Martin Pilhofer /   Abstract: Gamete formation and subsequent offspring development often involve extended phases of suspended cellular development or even dormancy. How cells adapt to recover and resume growth remains poorly ...Gamete formation and subsequent offspring development often involve extended phases of suspended cellular development or even dormancy. How cells adapt to recover and resume growth remains poorly understood. Here, we visualized budding yeast cells undergoing meiosis by cryo-electron tomography (cryoET) and discovered elaborate filamentous assemblies decorating the nucleus, cytoplasm, and mitochondria. To determine filament composition, we developed a "filament identification" (FilamentID) workflow that combines multiscale cryoET/cryo-electron microscopy (cryoEM) analyses of partially lysed cells or organelles. FilamentID identified the mitochondrial filaments as being composed of the conserved aldehyde dehydrogenase Ald4 and the nucleoplasmic/cytoplasmic filaments as consisting of acetyl-coenzyme A (CoA) synthetase Acs1. Structural characterization further revealed the mechanism underlying polymerization and enabled us to genetically perturb filament formation. Acs1 polymerization facilitates the recovery of chronologically aged spores and, more generally, the cell cycle re-entry of starved cells. FilamentID is broadly applicable to characterize filaments of unknown identity in diverse cellular contexts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19548.map.gz emd_19548.map.gz | 9.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19548-v30.xml emd-19548-v30.xml emd-19548.xml emd-19548.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19548.png emd_19548.png | 60.3 KB | ||
| Masks |  emd_19548_msk_1.map emd_19548_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19548.cif.gz emd-19548.cif.gz | 5.8 KB | ||
| Others |  emd_19548_additional_1.map.gz emd_19548_additional_1.map.gz emd_19548_half_map_1.map.gz emd_19548_half_map_1.map.gz emd_19548_half_map_2.map.gz emd_19548_half_map_2.map.gz | 46.9 MB 40.7 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19548 http://ftp.pdbj.org/pub/emdb/structures/EMD-19548 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19548 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19548 | HTTPS FTP |
-Validation report
| Summary document |  emd_19548_validation.pdf.gz emd_19548_validation.pdf.gz | 1002.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19548_full_validation.pdf.gz emd_19548_full_validation.pdf.gz | 1002.4 KB | Display | |
| Data in XML |  emd_19548_validation.xml.gz emd_19548_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_19548_validation.cif.gz emd_19548_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19548 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19548 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19548 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19548 | HTTPS FTP |
-Related structure data
| Related structure data |  8rwjMC  8rwkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19548.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19548.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19548_msk_1.map emd_19548_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: cryoEM map further improved by deepEMhancer
| File | emd_19548_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM map further improved by deepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19548_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19548_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Acetyl-CoA synthetase 1 filament from spread meiotic yeast sphero...
| Entire | Name: Acetyl-CoA synthetase 1 filament from spread meiotic yeast spheroplasts |
|---|---|
| Components |
|
-Supramolecule #1: Acetyl-CoA synthetase 1 filament from spread meiotic yeast sphero...
| Supramolecule | Name: Acetyl-CoA synthetase 1 filament from spread meiotic yeast spheroplasts type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Acetyl-coenzyme A synthetase
| Macromolecule | Name: Acetyl-coenzyme A synthetase / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO / EC number: acetate-CoA ligase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 79.27057 KDa |
| Sequence | String: MSPSAVQSSK LEEQSSEIDK LKAKMSQSAS TAQQKKEHEY EHLTSVKIVP QRPISDRLQP AIATHYSPHL DGLQDYQRLH KESIEDPAK FFGSKATQFL NWSKPFDKVF IPDSKTGRPS FQNNAWFLNG QLNACYNCVD RHALKTPNKK AIIFEGDEPG Q GYSITYKE ...String: MSPSAVQSSK LEEQSSEIDK LKAKMSQSAS TAQQKKEHEY EHLTSVKIVP QRPISDRLQP AIATHYSPHL DGLQDYQRLH KESIEDPAK FFGSKATQFL NWSKPFDKVF IPDSKTGRPS FQNNAWFLNG QLNACYNCVD RHALKTPNKK AIIFEGDEPG Q GYSITYKE LLEEVCQVAQ VLTYSMGVRK GDTVAVYMPM VPEAIITLLA ISRIGAIHSV VFAGFSSNSL RDRINDGDSK VV ITTDESN RGGKVIETKR IVDDALRETP GVRHVLVYRK TNNPSVAFHA PRDLDWATEK KKYKTYYPCT PVDSEDPLFL LYT SGSTGA PKGVQHSTAG YLLGALLTMR YTFDTHQEDV FFTAGDIGWI TGHTYVVYGP LLYGCATLVF EGTPAYPNYS RYWD IIDEH KVTQFYVAPT ALRLLKRAGD SYIENHSLKS LRCLGSVGEP IAAEVWEWYS EKIGKNEIPI VDTYWQTESG SHLVT PLAG GVTPMKPGSA SFPFFGIDAV VLDPNTGEEL NTSHAEGVLA VKAAWPSFAR TIWKNHDRYL DTYLNPYPGY YFTGDG AAK DKDGYIWILG RVDDVVNVSG HRLSTAEIEA AIIEDPIVAE CAVVGFNDDL TGQAVAAFVV LKNKSNWSTA TDDELQD IK KHLVFTVRKD IGPFAAPKLI ILVDDLPKTR SGKIMRRILR KILAGESDQL GDVSTLSNPG IVRHLIDSVK L UniProtKB: Acetyl-coenzyme A synthetase |
-Macromolecule #2: [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidany...
| Macromolecule | Name: [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate type: ligand / ID: 2 / Number of copies: 9 / Formula: 6R9 |
|---|---|
| Molecular weight | Theoretical: 389.258 Da |
| Chemical component information | 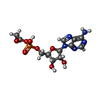 ChemComp-6R9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 53.61 Å Applied symmetry - Helical parameters - Δ&Phi: 13.03 ° Applied symmetry - Helical parameters - Axial symmetry: C3 (3 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 17169 |
|---|---|
| Startup model | Type of model: OTHER Details: the initial model is determined from sub-tomogram averaged volume of spread spheroplast. |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)