[English] 日本語
 Yorodumi
Yorodumi- EMDB-19551: cryo sub-tomogram average of Acs1 filament from spread meiotic ye... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 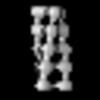 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo sub-tomogram average of Acs1 filament from spread meiotic yeast spheroplasts | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | metabolic enzyme / filament / cryoET / CYTOSOLIC PROTEIN / sub-tomogram average | |||||||||
| Biological species |   | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 18.3 Å | |||||||||
 Authors Authors | Hugener J / Xu J / Wettstein R / Ioannidi L / Velikov D / Wollweber F / Henggeler A / Matos J / Pilhofer M | |||||||||
| Funding support |  Switzerland, European Union, 2 items Switzerland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis. Authors: Jannik Hugener / Jingwei Xu / Rahel Wettstein / Lydia Ioannidi / Daniel Velikov / Florian Wollweber / Adrian Henggeler / Joao Matos / Martin Pilhofer /   Abstract: Gamete formation and subsequent offspring development often involve extended phases of suspended cellular development or even dormancy. How cells adapt to recover and resume growth remains poorly ...Gamete formation and subsequent offspring development often involve extended phases of suspended cellular development or even dormancy. How cells adapt to recover and resume growth remains poorly understood. Here, we visualized budding yeast cells undergoing meiosis by cryo-electron tomography (cryoET) and discovered elaborate filamentous assemblies decorating the nucleus, cytoplasm, and mitochondria. To determine filament composition, we developed a "filament identification" (FilamentID) workflow that combines multiscale cryoET/cryo-electron microscopy (cryoEM) analyses of partially lysed cells or organelles. FilamentID identified the mitochondrial filaments as being composed of the conserved aldehyde dehydrogenase Ald4 and the nucleoplasmic/cytoplasmic filaments as consisting of acetyl-coenzyme A (CoA) synthetase Acs1. Structural characterization further revealed the mechanism underlying polymerization and enabled us to genetically perturb filament formation. Acs1 polymerization facilitates the recovery of chronologically aged spores and, more generally, the cell cycle re-entry of starved cells. FilamentID is broadly applicable to characterize filaments of unknown identity in diverse cellular contexts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19551.map.gz emd_19551.map.gz | 120.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19551-v30.xml emd-19551-v30.xml emd-19551.xml emd-19551.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19551.png emd_19551.png | 15.1 KB | ||
| Filedesc metadata |  emd-19551.cif.gz emd-19551.cif.gz | 4.8 KB | ||
| Others |  emd_19551_half_map_1.map.gz emd_19551_half_map_1.map.gz emd_19551_half_map_2.map.gz emd_19551_half_map_2.map.gz | 120.3 KB 120.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19551 http://ftp.pdbj.org/pub/emdb/structures/EMD-19551 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19551 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19551 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19551.map.gz / Format: CCP4 / Size: 128.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19551.map.gz / Format: CCP4 / Size: 128.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 9.138 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19551_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
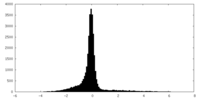
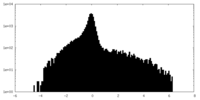
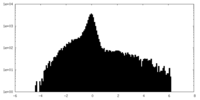
| Density Histograms |
-Half map: #2
| File | emd_19551_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
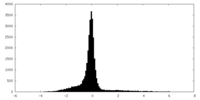
| Density Histograms |
- Sample components
Sample components
-Entire : Acetyl-CoA synthetase 1 polymer from spread meiotic yeast spheroplasts
| Entire | Name: Acetyl-CoA synthetase 1 polymer from spread meiotic yeast spheroplasts |
|---|---|
| Components |
|
-Supramolecule #1: Acetyl-CoA synthetase 1 polymer from spread meiotic yeast spheroplasts
| Supramolecule | Name: Acetyl-CoA synthetase 1 polymer from spread meiotic yeast spheroplasts type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Acetyl-CoA synthetase 1
| Macromolecule | Name: Acetyl-CoA synthetase 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSPSAVQSSK LEEQSSEID K LKAKMSQS AS TAQQKKE HEY EHLTSV KIVP QRPIS DRLQP AIAT HYSPHL DGL QDYQRLH KE SIEDPAKF F GSKATQFLN WSKPFDKVFI PDSKTGRPS F QNNAWFLN GQ LNACYNC VDR HALKTP NKKA IIFEG ...String: MSPSAVQSSK LEEQSSEID K LKAKMSQS AS TAQQKKE HEY EHLTSV KIVP QRPIS DRLQP AIAT HYSPHL DGL QDYQRLH KE SIEDPAKF F GSKATQFLN WSKPFDKVFI PDSKTGRPS F QNNAWFLN GQ LNACYNC VDR HALKTP NKKA IIFEG DEPGQ GYSI TYKELL EEV CQVAQVL TY SMGVRKGD T VAVYMPMVP EAIITLLAIS RIGAIHSVV F AGFSSNSL RD RINDGDS KVV ITTDES NRGG KVIET KRIVD DALR ETPGVR HVL VYRKTNN PS VAFHAPRD L DWATEKKKY KTYYPCTPVD SEDPLFLLY T SGSTGAPK GV QHSTAGY LLG ALLTMR YTFD THQED VFFTA GDIG WITGHT YVV YGPLLYG CA TLVFEGTP A YPNYSRYWD IIDEHKVTQF YVAPTALRL L KRAGDSYI EN HSLKSLR CLG SVGEPI AAEV WEWYS EKIGK NEIP IVDTYW QTE SGSHLVT PL AGGVTPMK P GSASFPFFG IDAVVLDPNT GEELNTSHA E GVLAVKAA WP SFARTIW KNH DRYLDT YLNP YPGYY FTGDG AAKD KDGYIW ILG RVDDVVN VS GHRLSTAE I EAAIIEDPI VAECAVVGFN DDLTGQAVA A FVVLKNKS NW STATDDE LQD IKKHLV FTVR KDIGP FAAPK LIIL VDDLPK TRS GKIMRRI LR KILAGESD Q LGDVSTLSN PGIVRHLIDS VKL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 2.13 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 8.0 µm / Nominal defocus min: 8.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 18.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number subtomograms used: 452 |
|---|---|
| Extraction | Number tomograms: 3 / Number images used: 452 |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller






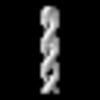
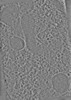




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)