+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | IFTA1 in retrograde Intraflagellar transport trains | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cilia / IFT / train / retrograde / PROTEIN TRANSPORT | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 16.6 Å | |||||||||
 Authors Authors | Lacey SE / Pigino G | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Extensive structural rearrangement of intraflagellar transport trains underpins bidirectional cargo transport. Authors: Samuel E Lacey / Andrea Graziadei / Gaia Pigino /  Abstract: Bidirectional transport in cilia is carried out by polymers of the IFTA and IFTB protein complexes, called anterograde and retrograde intraflagellar transport (IFT) trains. Anterograde trains deliver ...Bidirectional transport in cilia is carried out by polymers of the IFTA and IFTB protein complexes, called anterograde and retrograde intraflagellar transport (IFT) trains. Anterograde trains deliver cargoes from the cell to the cilium tip, then convert into retrograde trains for cargo export. We set out to understand how the IFT complexes can perform these two directly opposing roles before and after conversion. We use cryoelectron tomography and in situ cross-linking mass spectrometry to determine the structure of retrograde IFT trains and compare it with the known structure of anterograde trains. The retrograde train is a 2-fold symmetric polymer organized around a central thread of IFTA complexes. We conclude that anterograde-to-retrograde remodeling involves global rearrangements of the IFTA/B complexes and requires complete disassembly of the anterograde train. Finally, we describe how conformational changes to cargo-binding sites facilitate unidirectional cargo transport in a bidirectional system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19516.map.gz emd_19516.map.gz | 7.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19516-v30.xml emd-19516-v30.xml emd-19516.xml emd-19516.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19516.png emd_19516.png | 25.8 KB | ||
| Masks |  emd_19516_msk_1.map emd_19516_msk_1.map | 10.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19516.cif.gz emd-19516.cif.gz | 4.1 KB | ||
| Others |  emd_19516_additional_1.map.gz emd_19516_additional_1.map.gz emd_19516_half_map_1.map.gz emd_19516_half_map_1.map.gz emd_19516_half_map_2.map.gz emd_19516_half_map_2.map.gz | 68.5 KB 7.9 MB 7.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19516 http://ftp.pdbj.org/pub/emdb/structures/EMD-19516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19516 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19516.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19516.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.06 Å | ||||||||||||||||||||||||||||||||||||
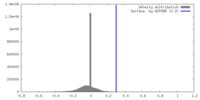
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19516_msk_1.map emd_19516_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
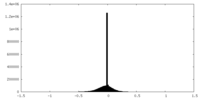
| Density Histograms |
-Additional map: The main map after Locspiral filtering and masking....
| File | emd_19516_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The main map after Locspiral filtering and masking. This was then used for composite map generation | ||||||||||||
| Projections & Slices |
| ||||||||||||
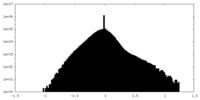
| Density Histograms |
-Half map: #1
| File | emd_19516_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19516_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : In situ retrograde intraflagellar transport trains in dhc1b-3 mut...
| Entire | Name: In situ retrograde intraflagellar transport trains in dhc1b-3 mutant Chlamydomonas reinhardtii cilia |
|---|---|
| Components |
|
-Supramolecule #1: In situ retrograde intraflagellar transport trains in dhc1b-3 mut...
| Supramolecule | Name: In situ retrograde intraflagellar transport trains in dhc1b-3 mutant Chlamydomonas reinhardtii cilia type: cell / ID: 1 / Parent: 0 Details: Temperature sensitive cells incubated at restrictive temperature (34 degrees C) for 10 hours and plunge frozen on grids. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 2.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 16.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1.3) / Number subtomograms used: 5896 |
|---|---|
| Extraction | Number tomograms: 756 / Number images used: 10455 |
| CTF correction | Software - Name: CTFFIND (ver. 4) / Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)