+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the undecorated barbed end of F-actin. | ||||||||||||
 Map data Map data | Sharpened, local-resolution filtered cryo-EM density map of the undecorated barbed end of actin filaments. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | actin / actin end / barbed end / actin assembly / STRUCTURAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament ...cytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / skeletal muscle fiber development / stress fiber / titin binding / actin filament polymerization / actin filament / filopodium / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / lamellipodium / cell body / protein domain specific binding / hydrolase activity / calcium ion binding / positive regulation of gene expression / magnesium ion binding / ATP binding / identical protein binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
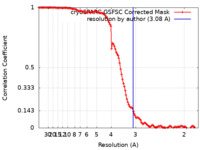
| Method | single particle reconstruction / cryo EM / Resolution: 3.08 Å | ||||||||||||
 Authors Authors | Oosterheert W / Boiero Sanders M / Funk J / Prumbaum D / Raunser S / Bieling P | ||||||||||||
| Funding support |  Germany, European Union, 3 items Germany, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Molecular mechanism of actin filament elongation by formins. Authors: Wout Oosterheert / Micaela Boiero Sanders / Johanna Funk / Daniel Prumbaum / Stefan Raunser / Peter Bieling /  Abstract: Formins control the assembly of actin filaments (F-actin) that drive cell morphogenesis and motility in eukaryotes. However, their molecular interaction with F-actin and their mechanism of action ...Formins control the assembly of actin filaments (F-actin) that drive cell morphogenesis and motility in eukaryotes. However, their molecular interaction with F-actin and their mechanism of action remain unclear. In this work, we present high-resolution cryo-electron microscopy structures of F-actin barbed ends bound by three distinct formins, revealing a common asymmetric formin conformation imposed by the filament. Formation of new intersubunit contacts during actin polymerization sterically displaces formin and triggers its translocation. This "undock-and-lock" mechanism explains how actin-filament growth is coordinated with formin movement. Filament elongation speeds are controlled by the positioning and stability of actin-formin interfaces, which distinguish fast and slow formins. Furthermore, we provide a structure of the actin-formin-profilin ring complex, which resolves how profilin is rapidly released from the barbed end during filament elongation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19501.map.gz emd_19501.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19501-v30.xml emd-19501-v30.xml emd-19501.xml emd-19501.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19501_fsc.xml emd_19501_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_19501.png emd_19501.png | 67.4 KB | ||
| Masks |  emd_19501_msk_1.map emd_19501_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19501.cif.gz emd-19501.cif.gz | 6.9 KB | ||
| Others |  emd_19501_additional_1.map.gz emd_19501_additional_1.map.gz emd_19501_half_map_1.map.gz emd_19501_half_map_1.map.gz emd_19501_half_map_2.map.gz emd_19501_half_map_2.map.gz | 89.3 MB 165.2 MB 165.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19501 http://ftp.pdbj.org/pub/emdb/structures/EMD-19501 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19501 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19501 | HTTPS FTP |
-Related structure data
| Related structure data |  8ru0MC  8rttC  8rtyC  8ru2C  8rv2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19501.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19501.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened, local-resolution filtered cryo-EM density map of the undecorated barbed end of actin filaments. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19501_msk_1.map emd_19501_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3D-refined, unsharpened cryo-EM density map of the undecorated...
| File | emd_19501_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D-refined, unsharpened cryo-EM density map of the undecorated barbed end of actin filaments. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map 2 of the undecorated barbed...
| File | emd_19501_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map 2 of the undecorated barbed end of actin filaments. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map 1 of the undecorated barbed...
| File | emd_19501_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map 1 of the undecorated barbed end of actin filaments. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the actin subunits that form the barbed end of actin f...
| Entire | Name: Complex of the actin subunits that form the barbed end of actin filaments. |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the actin subunits that form the barbed end of actin f...
| Supramolecule | Name: Complex of the actin subunits that form the barbed end of actin filaments. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Alpha actin was purified from rabbit skeletal muscle. Actin filaments were polymerized in the presence of the formin INF2, which was purified separetely and added during polymerization prior ...Details: Alpha actin was purified from rabbit skeletal muscle. Actin filaments were polymerized in the presence of the formin INF2, which was purified separetely and added during polymerization prior to cryo-EM grid preparation. |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Actin filament
| Supramolecule | Name: Actin filament / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 Details: Rabbit skeletal alpha actin purified from frozen rabbit muscle acetone powder. Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.875633 KDa |
| Sequence | String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IE(HIC)GII TNW DDMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSG DG VTHNVPIYEG ...String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IE(HIC)GII TNW DDMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSG DG VTHNVPIYEG YALPHAIMRL DLAGRDLTDY LMKILTERGY SFVTTAEREI VRDIKEKLCY VALDFENEMA TAASSSSL E KSYELPDGQV ITIGNERFRC PETLFQPSFI GMESAGIHET TYNSIMKCDI DIRKDLYANN VMSGGTTMYP GIADRMQKE ITALAPSTMK IKIIAPPERK YSVWIGGSIL ASLSTFQQMW ITKQEYDEAG PSIVHRKCF UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 4 / Number of copies: 3 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.1 Component:
Details: 12 mM HEPES pH 7.1, 100 mM KCl, 2.1 mM MgCl2, 1 mM EGTA, 1 mM TCEP, 0.2 mM ATP | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 286 K / Instrument: FEI VITROBOT MARK IV / Details: 3 seconds, force 0.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: 300 kV Titan Krios G3 microscope (Thermo Fisher Scientific). Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 15 eV / Details: Gatan energy filter. |
| Details | 300 kV Titan Krios G3 microscope (Thermo Fisher Scientific). |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 20305 / Average electron dose: 60.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)