+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Alpha-synuclein amyloid fibril | ||||||||||||
 Map data Map data | Postprocessed map of the alpha-synuclein amyloid fibril core | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Chaperones / amyloid fibril / PROTEIN FIBRIL | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of mitochondrial electron transport, NADH to ubiquinone / : / neutral lipid metabolic process / regulation of acyl-CoA biosynthetic process / negative regulation of dopamine uptake involved in synaptic transmission / negative regulation of norepinephrine uptake / response to desipramine / positive regulation of SNARE complex assembly / positive regulation of hydrogen peroxide catabolic process / supramolecular fiber ...negative regulation of mitochondrial electron transport, NADH to ubiquinone / : / neutral lipid metabolic process / regulation of acyl-CoA biosynthetic process / negative regulation of dopamine uptake involved in synaptic transmission / negative regulation of norepinephrine uptake / response to desipramine / positive regulation of SNARE complex assembly / positive regulation of hydrogen peroxide catabolic process / supramolecular fiber / regulation of synaptic vesicle recycling / negative regulation of chaperone-mediated autophagy / mitochondrial membrane organization / regulation of reactive oxygen species biosynthetic process / positive regulation of protein localization to cell periphery / negative regulation of platelet-derived growth factor receptor signaling pathway / negative regulation of exocytosis / regulation of glutamate secretion / dopamine biosynthetic process / response to iron(II) ion / SNARE complex assembly / negative regulation of dopamine metabolic process / positive regulation of neurotransmitter secretion / regulation of macrophage activation / positive regulation of inositol phosphate biosynthetic process / regulation of norepinephrine uptake / regulation of locomotion / synaptic vesicle transport / negative regulation of microtubule polymerization / transporter regulator activity / synaptic vesicle priming / dopamine uptake involved in synaptic transmission / protein kinase inhibitor activity / regulation of dopamine secretion / negative regulation of thrombin-activated receptor signaling pathway / dynein complex binding / mitochondrial ATP synthesis coupled electron transport / positive regulation of receptor recycling / cuprous ion binding / nuclear outer membrane / response to magnesium ion / positive regulation of exocytosis / synaptic vesicle exocytosis / positive regulation of endocytosis / kinesin binding / synaptic vesicle endocytosis / enzyme inhibitor activity / cysteine-type endopeptidase inhibitor activity / response to type II interferon / negative regulation of serotonin uptake / regulation of presynapse assembly / alpha-tubulin binding / beta-tubulin binding / phospholipase binding / behavioral response to cocaine / supramolecular fiber organization / cellular response to fibroblast growth factor stimulus / phospholipid metabolic process / axon terminus / inclusion body / cellular response to epinephrine stimulus / Hsp70 protein binding / response to interleukin-1 / regulation of microtubule cytoskeleton organization / cellular response to copper ion / positive regulation of release of sequestered calcium ion into cytosol / SNARE binding / adult locomotory behavior / excitatory postsynaptic potential / protein tetramerization / phosphoprotein binding / microglial cell activation / ferrous iron binding / fatty acid metabolic process / regulation of long-term neuronal synaptic plasticity / synapse organization / PKR-mediated signaling / protein destabilization / phospholipid binding / receptor internalization / tau protein binding / long-term synaptic potentiation / terminal bouton / positive regulation of inflammatory response / synaptic vesicle membrane / actin cytoskeleton / actin binding / growth cone / cellular response to oxidative stress / neuron apoptotic process / cell cortex / histone binding / response to lipopolysaccharide / microtubule binding / chemical synaptic transmission / amyloid fibril formation / molecular adaptor activity / negative regulation of neuron apoptotic process / oxidoreductase activity / mitochondrial outer membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Saibil HR / Monistrol J | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2025 Journal: Commun Biol / Year: 2025Title: Stepwise recruitment of chaperone Hsc70 by DNAJB1 produces ordered arrays primed for bursts of amyloid fibril disassembly. Authors: Jim Monistrol / Joseph G Beton / Erin C Johnston / Thi Lieu Dang / Bernd Bukau / Helen R Saibil /   Abstract: The Hsp70 chaperone system is capable of disassembling pathological aggregates such as amyloid fibres associated with serious degenerative diseases. Here we examine the role of the J-domain protein ...The Hsp70 chaperone system is capable of disassembling pathological aggregates such as amyloid fibres associated with serious degenerative diseases. Here we examine the role of the J-domain protein co-factor in amyloid disaggregation by the Hsc70 system. We used cryo-EM and tomography to compare the assemblies with wild-type DNAJB1 or inactive mutants. We show that DNAJB1 binds regularly along α-synuclein amyloid fibrils and acts in a 2-step recruitment of Hsc70, releasing DNAJB1 auto-inhibition before activating Hsc70 ATPase. The wild-type DNAJB1:Hsc70:Apg2 complex forms dense arrays of chaperones on the fibrils, with Hsc70 on the outer surface. When the auto-inhibition is removed by mutating DNAJB1 (ΔH5 DNAJB1), Hsc70 is recruited to the fibrils at a similar level, but the ΔH5 DNAJB1:Ηsc70:Apg2 complex is inactive, binds less regularly to the fibrils and lacks the ordered clusters. Therefore, we propose that 2-step activation of DNAJB1 regulates the ordered assembly of Hsc70 on the fibril. The localised, dense packing of chaperones could trigger a cascade of recruitment and activation to give coordinated, sequential binding and disaggregation from an exposed fibril end, as previously observed in AFM videos. This mechanism is likely to be important in maintaining a healthy cellular proteome into old age. #1:  Journal: Biorxiv / Year: 2024 Journal: Biorxiv / Year: 2024Title: Stepwise recruitment of Hsc70 by DNAJB1 produces ordered arrays primed for bursts of amyloid fibre disassembly Authors: Monistrol J / Beton JG / Johnston EC / Saibil HR | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19462.map.gz emd_19462.map.gz | 16.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19462-v30.xml emd-19462-v30.xml emd-19462.xml emd-19462.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19462.png emd_19462.png | 28 KB | ||
| Masks |  emd_19462_msk_1.map emd_19462_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19462.cif.gz emd-19462.cif.gz | 5.9 KB | ||
| Others |  emd_19462_half_map_1.map.gz emd_19462_half_map_1.map.gz emd_19462_half_map_2.map.gz emd_19462_half_map_2.map.gz | 96.9 MB 96.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19462 http://ftp.pdbj.org/pub/emdb/structures/EMD-19462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19462 | HTTPS FTP |
-Related structure data
| Related structure data |  8rrrMC  8rqmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19462.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19462.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map of the alpha-synuclein amyloid fibril core | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
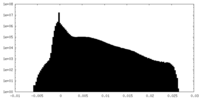
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19462_msk_1.map emd_19462_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
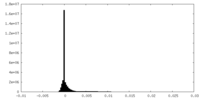
| Density Histograms |
-Half map: Unfiltered half map #2 of the alpha-synuclein amyloid fibril core
| File | emd_19462_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map #2 of the alpha-synuclein amyloid fibril core | ||||||||||||
| Projections & Slices |
| ||||||||||||
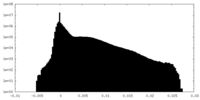
| Density Histograms |
-Half map: Unfiltered half map #1 of the alpha-synuclein amyloid fibril core
| File | emd_19462_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map #1 of the alpha-synuclein amyloid fibril core | ||||||||||||
| Projections & Slices |
| ||||||||||||
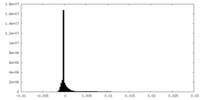
| Density Histograms |
- Sample components
Sample components
-Entire : Alpha-synuclein amyloid fibril
| Entire | Name: Alpha-synuclein amyloid fibril |
|---|---|
| Components |
|
-Supramolecule #1: Alpha-synuclein amyloid fibril
| Supramolecule | Name: Alpha-synuclein amyloid fibril / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Alpha-synuclein
| Macromolecule | Name: Alpha-synuclein / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.476108 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDVFMKGLSK AKEGVVAAAE KTKQGVAEAA GKTKEGVLYV GSKTKEGVVH GVATVAEKTK EQVTNVGGAV VTGVTAVAQK TVEGAGSIA AATGFVKKDQ LGKNEEGAPQ EGILEDMPVD PDNEAYEMPS EEGYQDYEPE A UniProtKB: Alpha-synuclein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 4.76 Å Applied symmetry - Helical parameters - Δ&Phi: -0.82 ° Applied symmetry - Helical parameters - Axial symmetry: C2 (2 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 245549 |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)