[English] 日本語
 Yorodumi
Yorodumi- EMDB-1928: Modular architecture of eukaryotic RNase P and RNase MRP revealed... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1928 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
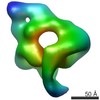
| Title | Modular architecture of eukaryotic RNase P and RNase MRP revealed by electron microscopy | |||||||||
 Map data Map data | RNase MRP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNase P / RNA-processing / ribozyme | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 15.0 Å | |||||||||
 Authors Authors | Hipp K / Galani K / Batisse C / Prinz S / Bottcher B | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2012 Journal: Nucleic Acids Res / Year: 2012Title: Modular architecture of eukaryotic RNase P and RNase MRP revealed by electron microscopy. Authors: Katharina Hipp / Kyriaki Galani / Claire Batisse / Simone Prinz / Bettina Böttcher /  Abstract: Ribonuclease P (RNase P) and RNase MRP are closely related ribonucleoprotein enzymes, which process RNA substrates including tRNA precursors for RNase P and 5.8 S rRNA precursors, as well as some ...Ribonuclease P (RNase P) and RNase MRP are closely related ribonucleoprotein enzymes, which process RNA substrates including tRNA precursors for RNase P and 5.8 S rRNA precursors, as well as some mRNAs, for RNase MRP. The structures of RNase P and RNase MRP have not yet been solved, so it is unclear how the proteins contribute to the structure of the complexes and how substrate specificity is determined. Using electron microscopy and image processing we show that eukaryotic RNase P and RNase MRP have a modular architecture, where proteins stabilize the RNA fold and contribute to cavities, channels and chambers between the modules. Such features are located at strategic positions for substrate recognition by shape and coordination of the cleaved-off sequence. These are also the sites of greatest difference between RNase P and RNase MRP, highlighting the importance of the adaptation of this region to the different substrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1928.map.gz emd_1928.map.gz | 3.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1928-v30.xml emd-1928-v30.xml emd-1928.xml emd-1928.xml | 9.1 KB 9.1 KB | Display Display |  EMDB header EMDB header |
| Images |  1928.png 1928.png | 82.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1928 http://ftp.pdbj.org/pub/emdb/structures/EMD-1928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1928 | HTTPS FTP |
-Validation report
| Summary document |  emd_1928_validation.pdf.gz emd_1928_validation.pdf.gz | 209.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1928_full_validation.pdf.gz emd_1928_full_validation.pdf.gz | 208.6 KB | Display | |
| Data in XML |  emd_1928_validation.xml.gz emd_1928_validation.xml.gz | 5.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1928 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1928 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1928 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1928 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1928.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1928.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RNase MRP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RNase MRP
| Entire | Name: RNase MRP |
|---|---|
| Components |
|
-Supramolecule #1000: RNase MRP
| Supramolecule | Name: RNase MRP / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 410 KDa |
-Macromolecule #1: Ribonuclease MRP
| Macromolecule | Name: Ribonuclease MRP / type: protein_or_peptide / ID: 1 / Name.synonym: RNase MRP / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 410 KDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 50 mM Tris HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT |
|---|---|
| Staining | Type: NEGATIVE Details: sandwich, cryo negative stain with 1% uranyl acetate |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: NITROGEN / Chamber temperature: 95 K / Instrument: OTHER Method: samples were stained and frozen after partly drying, by dipping grids into liquid nitrogen |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Min: 95 K / Max: 95 K / Average: 95 K |
| Alignment procedure | Legacy - Astigmatism: astigmatism was corrected at 200000 times magnification on carbon film |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS (2k x 2k) / Number real images: 2006 / Average electron dose: 20 e/Å2 / Bits/pixel: 12 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 1.1 µm / Nominal defocus min: 0.68 µm / Nominal magnification: 66000 |
| Sample stage | Specimen holder: side entry, liquid nitrogen cooled / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | particle were selected automatically, particle orientations were determined by projection matching |
|---|---|
| CTF correction | Details: each particle, phase flipping |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: spider, imagic, xmipp Details: final maps were calculated from 20 defocus groups, number of particles in certain orientations were limited to counterbalance preferred orientations Number images used: 32232 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















