[English] 日本語
 Yorodumi
Yorodumi- EMDB-19092: focused map of the peripheral arm of complex I from murine brain ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | focused map of the peripheral arm of complex I from murine brain in the closed conformation | |||||||||
 Map data Map data | focused map of the peripheral arm of complex I from murine brain in the closed conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | respiratory chain complex / mammalian mitochondria / MEMBRANE PROTEIN / ELECTRON TRANSPORT | |||||||||
| Biological species |  | |||||||||
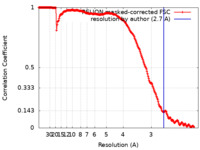
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Vercellino I / Sazanov LA | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: SCAF1 drives the compositional diversity of mammalian respirasomes. Authors: Irene Vercellino / Leonid A Sazanov /   Abstract: Supercomplexes of the respiratory chain are established constituents of the oxidative phosphorylation system, but their role in mammalian metabolism has been hotly debated. Although recent studies ...Supercomplexes of the respiratory chain are established constituents of the oxidative phosphorylation system, but their role in mammalian metabolism has been hotly debated. Although recent studies have shown that different tissues/organs are equipped with specific sets of supercomplexes, depending on their metabolic needs, the notion that supercomplexes have a role in the regulation of metabolism has been challenged. However, irrespective of the mechanistic conclusions, the composition of various high molecular weight supercomplexes remains uncertain. Here, using cryogenic electron microscopy, we demonstrate that mammalian (mouse) tissues contain three defined types of 'respirasome', supercomplexes made of CI, CIII and CIV. The stoichiometry and position of CIV differs in the three respirasomes, of which only one contains the supercomplex-associated factor SCAF1, whose involvement in respirasome formation has long been contended. Our structures confirm that the 'canonical' respirasome (the C-respirasome, CICIIICIV) does not contain SCAF1, which is instead associated to a different respirasome (the CS-respirasome), containing a second copy of CIV. We also identify an alternative respirasome (A-respirasome), with CIV bound to the 'back' of CI, instead of the 'toe'. This structural characterization of mouse mitochondrial supercomplexes allows us to hypothesize a mechanistic basis for their specific role in different metabolic conditions. #1:  Journal: Acta Crystallogr., Sect. D: Biol. Cristallogr. / Year: 2018 Journal: Acta Crystallogr., Sect. D: Biol. Cristallogr. / Year: 2018Title: Real-space refinement in PHENIX for cryo-EM and crystallography Authors: Afonine PV / Adams PD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19092.map.gz emd_19092.map.gz | 772.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19092-v30.xml emd-19092-v30.xml emd-19092.xml emd-19092.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19092_fsc.xml emd_19092_fsc.xml | 21 KB | Display |  FSC data file FSC data file |
| Images |  emd_19092.png emd_19092.png | 77.8 KB | ||
| Filedesc metadata |  emd-19092.cif.gz emd-19092.cif.gz | 4.6 KB | ||
| Others |  emd_19092_half_map_1.map.gz emd_19092_half_map_1.map.gz emd_19092_half_map_2.map.gz emd_19092_half_map_2.map.gz | 672.1 MB 673 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19092 http://ftp.pdbj.org/pub/emdb/structures/EMD-19092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19092 | HTTPS FTP |
-Validation report
| Summary document |  emd_19092_validation.pdf.gz emd_19092_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19092_full_validation.pdf.gz emd_19092_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_19092_validation.xml.gz emd_19092_validation.xml.gz | 28.9 KB | Display | |
| Data in CIF |  emd_19092_validation.cif.gz emd_19092_validation.cif.gz | 38.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19092 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19092 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19092 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19092 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19092.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19092.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused map of the peripheral arm of complex I from murine brain in the closed conformation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map 1
| File | emd_19092_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_19092_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Closed Complex I from murine brain
| Entire | Name: Closed Complex I from murine brain |
|---|---|
| Components |
|
-Supramolecule #1: Closed Complex I from murine brain
| Supramolecule | Name: Closed Complex I from murine brain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#44 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.7 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 10416 / Average exposure time: 4.4 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.6 µm / Calibrated defocus min: 0.6 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | initial fitting done in chimera |
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller












































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)