[English] 日本語
 Yorodumi
Yorodumi- EMDB-18944: Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroup... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - E particle | |||||||||
 Map data Map data | Main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Enterovirus / coxsackievirus / thermostable / mutant / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Coxsackievirus B5 Coxsackievirus B5 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Kumar K / Antanasijevic A | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Environ Sci Technol / Year: 2024 Journal: Environ Sci Technol / Year: 2024Title: Influence of Amino Acid Substitutions in Capsid Proteins of Coxsackievirus B5 on Free Chlorine and Thermal Inactivation. Authors: Shotaro Torii / Jérôme Gouttenoire / Kiruthika Kumar / Aleksandar Antanasijevic / Tamar Kohn /  Abstract: The sensitivity of enteroviruses to disinfectants varies among genetically similar variants and coincides with amino acid changes in capsid proteins, although the effect of individual substitutions ...The sensitivity of enteroviruses to disinfectants varies among genetically similar variants and coincides with amino acid changes in capsid proteins, although the effect of individual substitutions remains unknown. Here, we employed reverse genetics to investigate how amino acid substitutions in coxsackievirus B5 (CVB5) capsid proteins affect the virus' sensitivity to free chlorine and heat treatment. Of ten amino acid changes observed in CVB5 variants with free chlorine resistance, none significantly reduced the chlorine sensitivity, indicating a minor role of the capsid composition in chlorine sensitivity of CVB5. Conversely, a subset of these amino acid changes located at the C-terminal region of viral protein 1 led to reduced heat sensitivity. Cryo-electron microscopy revealed that these changes affect the assembly of intermediate viral states (altered and empty particles), suggesting that the mechanism for reduced heat sensitivity could be related to improved molecular packing of CVB5, resulting in greater stability or altered dynamics of virus uncoating during infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18944.map.gz emd_18944.map.gz | 207.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18944-v30.xml emd-18944-v30.xml emd-18944.xml emd-18944.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18944_fsc.xml emd_18944_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18944.png emd_18944.png | 230.7 KB | ||
| Masks |  emd_18944_msk_1.map emd_18944_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18944.cif.gz emd-18944.cif.gz | 6.7 KB | ||
| Others |  emd_18944_half_map_1.map.gz emd_18944_half_map_1.map.gz emd_18944_half_map_2.map.gz emd_18944_half_map_2.map.gz | 391.7 MB 391.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18944 http://ftp.pdbj.org/pub/emdb/structures/EMD-18944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18944 | HTTPS FTP |
-Validation report
| Summary document |  emd_18944_validation.pdf.gz emd_18944_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18944_full_validation.pdf.gz emd_18944_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_18944_validation.xml.gz emd_18944_validation.xml.gz | 24.9 KB | Display | |
| Data in CIF |  emd_18944_validation.cif.gz emd_18944_validation.cif.gz | 32.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18944 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18944 | HTTPS FTP |
-Related structure data
| Related structure data |  8r5zMC  8r5xC  8r5yC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18944.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18944.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.926 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18944_msk_1.map emd_18944_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
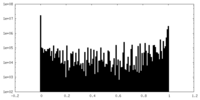
| Density Histograms |
-Half map: half map 1
| File | emd_18944_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
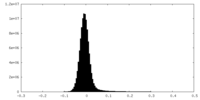
| Density Histograms |
-Half map: half map 2
| File | emd_18944_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Coxsackievirus B5
| Entire | Name:  Coxsackievirus B5 Coxsackievirus B5 |
|---|---|
| Components |
|
-Supramolecule #1: Coxsackievirus B5
| Supramolecule | Name: Coxsackievirus B5 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Particle E state of the CVB5F.cas.genogroupB mutant. NCBI-ID: 12074 / Sci species name: Coxsackievirus B5 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: Capsid VP1-3 / Diameter: 300.0 Å |
-Macromolecule #1: Genome polyprotein
| Macromolecule | Name: Genome polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B5 Coxsackievirus B5 |
| Molecular weight | Theoretical: 93.795359 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAQVSTQKT GAHETGLNAS GNSIIHYTNM NYYKDSASNS ANRQEFAQDP GKFTEPVKDI MIKSMPALNS PSAEECGYSD RVRSITLGN STITTQECAN VVVGYGTWPT YLRDEEATAE DQPTQPDVAT CRFYTLESVM WQQSSPGWWW KFPDALSNMG L FGQNMQYH ...String: MGAQVSTQKT GAHETGLNAS GNSIIHYTNM NYYKDSASNS ANRQEFAQDP GKFTEPVKDI MIKSMPALNS PSAEECGYSD RVRSITLGN STITTQECAN VVVGYGTWPT YLRDEEATAE DQPTQPDVAT CRFYTLESVM WQQSSPGWWW KFPDALSNMG L FGQNMQYH YLGRAGYTLH VQCNASKFHQ GCLLVVCVPE AEMGCATIAN KPDQKSLSNG ETANVFDSQN TSGQTAVQAN VI NAGMGIG VGNLTIFPHQ WINLRTNNSA TIVMPYVNSV PMDNMFRHNN FTLMIIPFAP LSYSTGATTY VPITVTVAPM CAE YNGLRL AGRQGLPTML TPGSNQFLTS DDFQSPSAMP QFDVTPEMAI PGQVNNLMEI AEVDSVVPVN NTEGKVMSIE AYQI PVQSN STNGSQVFGF PLIPGASSVL NRTLLGEILN YYTHWSGSIK LTFMFCGSAM ATGKFLLAYS PPGAGAPTTR KEAML GTHV IWDVGLQSSC VLCIPWISQT HYRYVVMDEY TAGGYITCWY QTNIVVPADT QSDCKILCFV SACNDFSVRM LKDTPF IKQ DSFFQGPPGE AIERAIARVA DTISSGPVNS ESIPALTAAE TGHTSQVVPA DTMQTRHVKN YHSRSESTVE NFLCRSA CV YYTTYKNHGT DGDNFAQWVI NTRQVAQLRR KLEMFTYARF DLELTFVITS SQEQSTIKGQ DSPVLTHQIM YVPPGGPV P TKINSYSWQT STNPSVFWTE GSAPPRISIP FISIGNAYSM FYDGWARFDK QGTYGINTLN NMGTLYMRHV NDGSPGPIV STVRIYFKPK HVKTWVPRPP RLCQYQKAGN VNFEPSGVTE GRTEITAMQT T UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: TBS | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV Details: 3ul of sample applied. Blotting time varied. Blotting force = 0. Total blots = 1.. | |||||||||
| Details | Inactivated by formaldehyde. Purified using a combination of sucrose gradient and size-exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 2 / Number real images: 6653 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model Details: Asymmetric unit used for fitting and interpretation |
|---|---|
| Details | Initial fitting was performed in Chimera and model refinement was performed in Coot and Rosetta. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8r5z: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)