[English] 日本語
 Yorodumi
Yorodumi- EMDB-18541: Structure of the mu opioid receptor bound to the antagonist nanob... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the mu opioid receptor bound to the antagonist nanobody NbE | ||||||||||||
 Map data Map data | DeepEMhancer sharpened map of the MOR_NbE_NabFab_anti-Fab complex. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | opioid receptor / nanobody / antagonist / NabFab / MOR / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationOpioid Signalling / Peptide ligand-binding receptors / beta-endorphin receptor activity / morphine receptor activity / negative regulation of Wnt protein secretion / G-protein activation / G protein-coupled opioid receptor activity / G protein-coupled opioid receptor signaling pathway / G alpha (i) signalling events / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway ...Opioid Signalling / Peptide ligand-binding receptors / beta-endorphin receptor activity / morphine receptor activity / negative regulation of Wnt protein secretion / G-protein activation / G protein-coupled opioid receptor activity / G protein-coupled opioid receptor signaling pathway / G alpha (i) signalling events / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / negative regulation of nitric oxide biosynthetic process / regulation of NMDA receptor activity / positive regulation of neurogenesis / negative regulation of cytosolic calcium ion concentration / transmission of nerve impulse / G-protein alpha-subunit binding / sensory perception of pain / presynaptic modulation of chemical synaptic transmission / locomotory behavior / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / adenylate cyclase-activating dopamine receptor signaling pathway / presynapse / perikaryon / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / endosome / axon / dendrite / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Yu J / Kumar A / Zhang X / Martin C / Raia P / Manglik A / Ballet S / Boland A / Stoeber M | ||||||||||||
| Funding support |  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation | Journal: bioRxiv / Year: 2023 Title: Structural Basis of μ-Opioid Receptor-Targeting by a Nanobody Antagonist. Authors: Jun Yu / Amit Kumar / Xuefeng Zhang / Charlotte Martin / Pierre Raia / Antoine Koehl / Toon Laeremans / Jan Steyaert / Aashish Manglik / Steven Ballet / Andreas Boland / Miriam Stoeber /    Abstract: The μ-opioid receptor (μOR), a prototypical member of the G protein-coupled receptor (GPCR) family, is the molecular target of opioid analgesics such as morphine and fentanyl. Due to the ...The μ-opioid receptor (μOR), a prototypical member of the G protein-coupled receptor (GPCR) family, is the molecular target of opioid analgesics such as morphine and fentanyl. Due to the limitations and severe side effects of currently available opioid drugs, there is considerable interest in developing novel modulators of μOR function. Most GPCR ligands today are small molecules, however biologics, including antibodies and nanobodies, are emerging as alternative therapeutics with clear advantages such as affinity and target selectivity. Here, we describe the nanobody NbE, which selectively binds to the μOR and acts as an antagonist. We functionally characterize NbE as an extracellular and genetically encoded μOR ligand and uncover the molecular basis for μOR antagonism by solving the cryo-EM structure of the NbE-μOR complex. NbE displays a unique ligand binding mode and achieves μOR selectivity by interactions with the orthosteric pocket and extracellular receptor loops. Based on a β-hairpin loop formed by NbE that deeply inserts into the μOR and centers most binding contacts, we design short peptide analogues that retain μOR antagonism. The work illustrates the potential of nanobodies to uniquely engage with GPCRs and describes novel μOR ligands that can serve as a basis for therapeutic developments. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18541.map.gz emd_18541.map.gz | 108.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18541-v30.xml emd-18541-v30.xml emd-18541.xml emd-18541.xml | 38.1 KB 38.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18541.png emd_18541.png | 29.5 KB | ||
| Masks |  emd_18541_msk_1.map emd_18541_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18541.cif.gz emd-18541.cif.gz | 8.1 KB | ||
| Others |  emd_18541_additional_1.map.gz emd_18541_additional_1.map.gz emd_18541_additional_2.map.gz emd_18541_additional_2.map.gz emd_18541_additional_3.map.gz emd_18541_additional_3.map.gz emd_18541_additional_4.map.gz emd_18541_additional_4.map.gz emd_18541_additional_5.map.gz emd_18541_additional_5.map.gz emd_18541_half_map_1.map.gz emd_18541_half_map_1.map.gz emd_18541_half_map_2.map.gz emd_18541_half_map_2.map.gz | 62.8 MB 62.9 MB 109.4 MB 115.9 MB 115.9 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18541 http://ftp.pdbj.org/pub/emdb/structures/EMD-18541 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18541 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18541 | HTTPS FTP |
-Validation report
| Summary document |  emd_18541_validation.pdf.gz emd_18541_validation.pdf.gz | 685.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18541_full_validation.pdf.gz emd_18541_full_validation.pdf.gz | 685.2 KB | Display | |
| Data in XML |  emd_18541_validation.xml.gz emd_18541_validation.xml.gz | 13.7 KB | Display | |
| Data in CIF |  emd_18541_validation.cif.gz emd_18541_validation.cif.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18541 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18541 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18541 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18541 | HTTPS FTP |
-Related structure data
| Related structure data |  8qotMC  8v8kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18541.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18541.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map of the MOR_NbE_NabFab_anti-Fab complex. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9759 Å | ||||||||||||||||||||||||||||||||||||
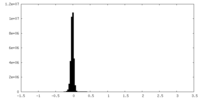
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18541_msk_1.map emd_18541_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
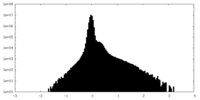
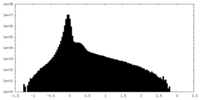
| Density Histograms |
-Additional map: Unsharpened map of the MOR NbE NabFab anti-Fab complex.
| File | emd_18541_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of the MOR_NbE_NabFab_anti-Fab complex. | ||||||||||||
| Projections & Slices |
| ||||||||||||
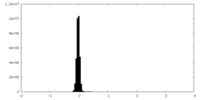
| Density Histograms |
-Additional map: Unsharpened MOR NbE map (focused refinement).
| File | emd_18541_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened MOR_NbE map (focused refinement). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: DeepEMhancer sharpened MOR NbE map (focused refinement).
| File | emd_18541_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened MOR_NbE map (focused refinement). | ||||||||||||
| Projections & Slices |
| ||||||||||||
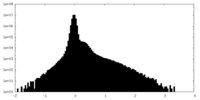
| Density Histograms |
-Additional map: Half map A of the MOR NbE map (focused refinement).
| File | emd_18541_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of the MOR_NbE map (focused refinement). | ||||||||||||
| Projections & Slices |
| ||||||||||||
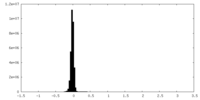
| Density Histograms |
-Additional map: Half map B of the MOR NbE map (focused refinement).
| File | emd_18541_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of the MOR_NbE map (focused refinement). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A of the MOR NbE NabFab anti-Fab complex.
| File | emd_18541_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of the MOR_NbE_NabFab_anti-Fab complex. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of the MOR NbE NabFab anti-Fab complex.
| File | emd_18541_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of the MOR_NbE_NabFab_anti-Fab complex. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MOR-NbE-NabFab-AntiFab complex
| Entire | Name: MOR-NbE-NabFab-AntiFab complex |
|---|---|
| Components |
|
-Supramolecule #1: MOR-NbE-NabFab-AntiFab complex
| Supramolecule | Name: MOR-NbE-NabFab-AntiFab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Mu-opioid receptor bound to a nanobody antagonist (NbE), and NabFab module used as fiducial marker. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Mu-type opioid receptor
| Macromolecule | Name: Mu-type opioid receptor / type: protein_or_peptide / ID: 1 / Details: MOR-2xSTREP-8xHIS / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.994215 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAMGPGNI SDCSDPLAPA SCSPAPGSWL NLSHVDGNQS DPCGPNRTGL GENLYFQGSH SLCPQTGSP SMVTAITIMA LYSIVCVVGL FGNFLVMYVI VRYTKMKTAT NIYIFNLALA DALATSTLPF QSVNYLMGTW P FGNILCKI ...String: MKTIIALSYI FCLVFADYKD DDDAMGPGNI SDCSDPLAPA SCSPAPGSWL NLSHVDGNQS DPCGPNRTGL GENLYFQGSH SLCPQTGSP SMVTAITIMA LYSIVCVVGL FGNFLVMYVI VRYTKMKTAT NIYIFNLALA DALATSTLPF QSVNYLMGTW P FGNILCKI VISIDYYNMF TSIFTLCTMS VDRYIAVCHP VKALDFRTPR NAKIVNVCNW ILSSAIGLPV MFMATTKYRQ GS IDCTLTF SHPTWYWENL LKICVFIFAF IMPVLIITVC YGLMILRLKS VRMLSGSKEK DRNLRRITRM VLVVVAVFIV CWT PIHIYV IIKALITIPE TTFQTVSWHF CIALGYTNSC LNPVLYAFLD ENFKRCFREF CIPTSSTILE VLFQGPEQQN SARI RQNTR EHPSTANTVD RTNHQLENLE AETAPLPSSL AEENLYFQSW SHPQFEKGGG SGGGSGGGSW SHPQFEKGAH HHHHH HH UniProtKB: Mu-type opioid receptor |
-Macromolecule #2: Nanobody E (NbE)
| Macromolecule | Name: Nanobody E (NbE) / type: protein_or_peptide / ID: 2 / Details: synthesized nanobody NbE-6xHis / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.458662 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VKKLLFAIPL VVPFYAAQPA MAQVQLVESG GGLVQAGGSL RLSCAASGSI SSISTMGWYR QAPGKERELV AAITSGGSTN YADSVKGRF TISRDNAKNT VYLQMNSLKP EDTAVYYCNF KYYSGSYFYK SEYDYWGKGT PVTVSSAAAH HHHHHGAAEQ K LISEEDLN GAA |
-Macromolecule #3: NabFab HC
| Macromolecule | Name: NabFab HC / type: protein_or_peptide / ID: 3 / Details: Signal sequence-NabFab HC / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 28.220525 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKNIAFLLA SMFVFSIATN AYAEISEVQL VESGGGLVQP GGSLRLSCAA SGFNFSYYSI HWVRQAPGKG LEWVAYISSS SSYTSYADS VKGRFTISAD TSKNTAYLQM NSLRAEDTAV YYCARGYQYW QYHASWYWNG GLDYWGQGTL VTVSSASTKG P SVFPLAPS ...String: MKKNIAFLLA SMFVFSIATN AYAEISEVQL VESGGGLVQP GGSLRLSCAA SGFNFSYYSI HWVRQAPGKG LEWVAYISSS SSYTSYADS VKGRFTISAD TSKNTAYLQM NSLRAEDTAV YYCARGYQYW QYHASWYWNG GLDYWGQGTL VTVSSASTKG P SVFPLAPS SKSTSGGTAA LGCLVKDYFP EPVTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VN HKPSNTK VDKKVEPKSC DKTHT |
-Macromolecule #4: NabFab LC
| Macromolecule | Name: NabFab LC / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 25.794859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKNIAFLLA SMFVFSIATN AYASDIQMTQ SPSSLSASVG DRVTITCRAS QSVSSAVAWY QQKPGKAPKL LIYSASSLYS GVPSRFSGS RSGTDFTLTI SSLQPEDFAT YYCQQSSSSL ITFGQGTKVE IKRTVAAPSV FIFPPSDSQL KSGTASVVCL L NNFYPREA ...String: MKKNIAFLLA SMFVFSIATN AYASDIQMTQ SPSSLSASVG DRVTITCRAS QSVSSAVAWY QQKPGKAPKL LIYSASSLYS GVPSRFSGS RSGTDFTLTI SSLQPEDFAT YYCQQSSSSL ITFGQGTKVE IKRTVAAPSV FIFPPSDSQL KSGTASVVCL L NNFYPREA KVQWKVDNAL QSGNSQESVT EQDSKDSTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #5: Anti-Fab Nanobody
| Macromolecule | Name: Anti-Fab Nanobody / type: protein_or_peptide / ID: 5 / Details: Signal sequence-6xHis Anti-Fab nanobody / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.414383 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAMHHHHHHG ENLYFQGSQV QLQESGGGLV QPGGSLRLSC AASGRTISRY AMSWFRQAPG KEREFVAVA RRSGDGAFYA DSVQGRFTVS RDDAKNTVYL QMNSLKPEDT AVYYCAIDSD TFYSGSYDYW GQGTQVTVSS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.8 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 288 K / Instrument: LEICA EM GP | |||||||||||||||
| Details | MOR receptor was expressed and purified in insect cells in isolation. NbE was expressed and purified in E. coli. NabFab module was expressed and purified in E. coli. All purified components were incubated and formed readily a stable complex. Final complex was purified over size exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 90.0 K / Max: 95.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 4 / Number real images: 5938 / Average exposure time: 52.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 68 | ||||||||||
| Output model |  PDB-8qot: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)