+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1832 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
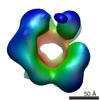
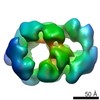
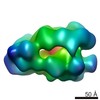
| Title | Drosophila melanogaster CMG complex bound to ADP.BeF3 | |||||||||
 Map data Map data | Volume file of the ADP.BeF3-bound CMG complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ ATPase / Helicase / DNA replication | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / Resolution: 28.0 Å | |||||||||
 Authors Authors | Costa A / Ilves I / Tamberg N / Petojevic T / Nogales E / Botchan MR / Berger JM | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2011 Journal: Nat Struct Mol Biol / Year: 2011Title: The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Authors: Alessandro Costa / Ivar Ilves / Nele Tamberg / Tatjana Petojevic / Eva Nogales / Michael R Botchan / James M Berger /  Abstract: Two central steps for initiating eukaryotic DNA replication involve loading of the Mcm2-7 helicase onto double-stranded DNA and its activation by GINS-Cdc45. To better understand these events, we ...Two central steps for initiating eukaryotic DNA replication involve loading of the Mcm2-7 helicase onto double-stranded DNA and its activation by GINS-Cdc45. To better understand these events, we determined the structures of Mcm2-7 and the CMG complex by using single-particle electron microscopy. Mcm2-7 adopts two conformations--a lock-washer-shaped spiral state and a planar, gapped-ring form--in which Mcm2 and Mcm5 flank a breach in the helicase perimeter. GINS and Cdc45 bridge this gap, forming a topologically closed assembly with a large interior channel; nucleotide binding further seals off the discontinuity between Mcm2 and Mcm5, partitioning the channel into two smaller pores. Together, our data help explain how GINS and Cdc45 activate Mcm2-7, indicate that Mcm2-7 loading may be assisted by a natural predisposition of the hexamer to form open rings, and suggest a mechanism by which the CMG complex assists DNA strand separation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1832.map.gz emd_1832.map.gz | 348.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1832-v30.xml emd-1832-v30.xml emd-1832.xml emd-1832.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_1832.png emd_1832.png | 135.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1832 http://ftp.pdbj.org/pub/emdb/structures/EMD-1832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1832 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1832.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1832.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Volume file of the ADP.BeF3-bound CMG complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Drosophila melanogaster CMG complex bound to ADP.BeF3
+Supramolecule #1000: Drosophila melanogaster CMG complex bound to ADP.BeF3
+Macromolecule #1: Mcm2
+Macromolecule #2: Mcm3
+Macromolecule #3: Mcm4
+Macromolecule #4: Mcm5
+Macromolecule #5: Mcm6
+Macromolecule #6: Mcm7
+Macromolecule #7: Psf1
+Macromolecule #8: Psf2
+Macromolecule #9: Psf3
+Macromolecule #10: Sld5
+Macromolecule #11: Cdc45
-Experimental details
-Structure determination
 Processing Processing | single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Average electron dose: 20 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 30000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 28.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Spider |
|---|
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















