[English] 日本語
 Yorodumi
Yorodumi- EMDB-18318: Cryo-EM structure of Vipp1 helical filament with lattice 1 (Vipp1_L1) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Vipp1 helical filament with lattice 1 (Vipp1_L1) | |||||||||
 Map data Map data | Primary sharpened map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Vipp1/IM30/ESCRT-III / Membrane remodeling / Cryoelectron microscopy / Helical filament structure / LIPID BINDING PROTEIN | |||||||||
| Function / homology | PspA/IM30 / PspA/IM30 family / lipid binding / plasma membrane / Membrane-associated protein Vipp1 Function and homology information Function and homology information | |||||||||
| Biological species |  Nostoc punctiforme (bacteria) Nostoc punctiforme (bacteria) | |||||||||
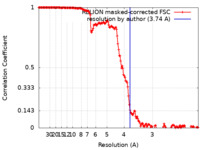
| Method | helical reconstruction / cryo EM / Resolution: 3.74 Å | |||||||||
 Authors Authors | Naskar S / Low HH | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Mechanism for Vipp1 spiral formation, ring biogenesis, and membrane repair. Authors: Souvik Naskar / Andrea Merino / Javier Espadas / Jayanti Singh / Aurelien Roux / Adai Colom / Harry H Low /    Abstract: The ESCRT-III-like protein Vipp1 couples filament polymerization with membrane remodeling. It assembles planar sheets as well as 3D rings and helical polymers, all implicated in mitigating plastid- ...The ESCRT-III-like protein Vipp1 couples filament polymerization with membrane remodeling. It assembles planar sheets as well as 3D rings and helical polymers, all implicated in mitigating plastid-associated membrane stress. The architecture of Vipp1 planar sheets and helical polymers remains unknown, as do the geometric changes required to transition between polymeric forms. Here we show how cyanobacterial Vipp1 assembles into morphologically-related sheets and spirals on membranes in vitro. The spirals converge to form a central ring similar to those described in membrane budding. Cryo-EM structures of helical filaments reveal a close geometric relationship between Vipp1 helical and planar lattices. Moreover, the helical structures reveal how filaments twist-a process required for Vipp1, and likely other ESCRT-III filaments, to transition between planar and 3D architectures. Overall, our results provide a molecular model for Vipp1 ring biogenesis and a mechanism for Vipp1 membrane stabilization and repair, with implications for other ESCRT-III systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18318.map.gz emd_18318.map.gz | 306.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18318-v30.xml emd-18318-v30.xml emd-18318.xml emd-18318.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18318_fsc.xml emd_18318_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18318.png emd_18318.png | 109.8 KB | ||
| Filedesc metadata |  emd-18318.cif.gz emd-18318.cif.gz | 6.2 KB | ||
| Others |  emd_18318_additional_1.map.gz emd_18318_additional_1.map.gz emd_18318_additional_2.map.gz emd_18318_additional_2.map.gz emd_18318_half_map_1.map.gz emd_18318_half_map_1.map.gz emd_18318_half_map_2.map.gz emd_18318_half_map_2.map.gz | 273.7 MB 324.7 MB 276.1 MB 276.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18318 http://ftp.pdbj.org/pub/emdb/structures/EMD-18318 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18318 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18318 | HTTPS FTP |
-Related structure data
| Related structure data |  8qbrMC  8qbsC  8qbvC  8qbwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18318.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18318.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary sharpened map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||
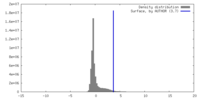
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened and unmasked raw map from 3D auto-refinement.
| File | emd_18318_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened and unmasked raw map from 3D auto-refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
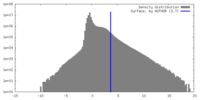
| Density Histograms |
-Additional map: Additional sharpened map produced by DeepEMhancer.
| File | emd_18318_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional sharpened map produced by DeepEMhancer. | ||||||||||||
| Projections & Slices |
| ||||||||||||
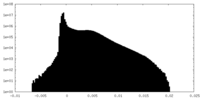
| Density Histograms |
-Half map: Unmasked raw half-map 1.
| File | emd_18318_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked raw half-map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
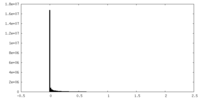
| Density Histograms |
-Half map: Unmasked raw half-map 2.
| File | emd_18318_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked raw half-map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Vipp1_L1
| Entire | Name: Vipp1_L1 |
|---|---|
| Components |
|
-Supramolecule #1: Vipp1_L1
| Supramolecule | Name: Vipp1_L1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Nostoc punctiforme (bacteria) Nostoc punctiforme (bacteria) |
-Macromolecule #1: Membrane-associated protein Vipp1
| Macromolecule | Name: Membrane-associated protein Vipp1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc punctiforme (bacteria) Nostoc punctiforme (bacteria) |
| Molecular weight | Theoretical: 23.9621 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGLFDRIKRV VSSNLNDLVN KAEDPEKMLE QAILEMQEDL VQLRQGVAQA IAAQKRSEKQ YNDAQNEINK WQRNAQLALQ KGDENLARQ ALERKKTYTD TSAALKASLD TQSTQVETLK RNLIQLESKI SEAKTKKEML KARITTAKAQ EQLQGMVRGM N TSSAMSAF ...String: MGLFDRIKRV VSSNLNDLVN KAEDPEKMLE QAILEMQEDL VQLRQGVAQA IAAQKRSEKQ YNDAQNEINK WQRNAQLALQ KGDENLARQ ALERKKTYTD TSAALKASLD TQSTQVETLK RNLIQLESKI SEAKTKKEML KARITTAKAQ EQLQGMVRGM N TSSAMSAF ERMEEKVLMQ ESRAQALGEL AGADLETQFA QLEGGSDVDD ELAALK UniProtKB: Membrane-associated protein Vipp1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1.7 mg/mL |
|---|---|
| Buffer | pH: 8.4 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 50 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.02 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Overall B value: 91.05 |
|---|---|
| Output model |  PDB-8qbr: |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)