+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ATP-bound IstB in complex to duplex DNA | |||||||||
 Map data Map data | Sharpened Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA Transposition / transposon / transpososome / AAA+ ATPases / target DNA / IS21 / IstB / Insertion sequence / DNA BINDING PROTEIN / cryo-electron microscopy. | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) | |||||||||
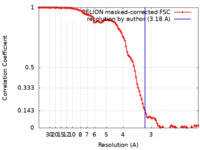
| Method | single particle reconstruction / cryo EM / Resolution: 3.18 Å | |||||||||
 Authors Authors | de la Gandara A / Spinola-Amilibia M / Araujo-Bazan L / Nunez-Ramirez R / Berger JM / Arias-Palomo E | |||||||||
| Funding support |  Spain, 2 items Spain, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Molecular basis for transposase activation by a dedicated AAA+ ATPase. Authors: Álvaro de la Gándara / Mercedes Spínola-Amilibia / Lidia Araújo-Bazán / Rafael Núñez-Ramírez / James M Berger / Ernesto Arias-Palomo /   Abstract: Transposases drive chromosomal rearrangements and the dissemination of drug-resistance genes and toxins. Although some transposases act alone, many rely on dedicated AAA+ ATPase subunits that ...Transposases drive chromosomal rearrangements and the dissemination of drug-resistance genes and toxins. Although some transposases act alone, many rely on dedicated AAA+ ATPase subunits that regulate site selectivity and catalytic function through poorly understood mechanisms. Using IS21 as a model transposase system, we show how an ATPase regulator uses nucleotide-controlled assembly and DNA deformation to enable structure-based site selectivity, transposase recruitment, and activation and integration. Solution and cryogenic electron microscopy studies show that the IstB ATPase self-assembles into an autoinhibited pentamer of dimers that tightly curves target DNA into a half-coil. Two of these decamers dimerize, which stabilizes the target nucleic acid into a kinked S-shaped configuration that engages the IstA transposase at the interface between the two IstB oligomers to form an approximately 1 MDa transpososome complex. Specific interactions stimulate regulator ATPase activity and trigger a large conformational change on the transposase that positions the catalytic site to perform DNA strand transfer. These studies help explain how AAA+ ATPase regulators-which are used by classical transposition systems such as Tn7, Mu and CRISPR-associated elements-can remodel their substrate DNA and cognate transposases to promote function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18136.map.gz emd_18136.map.gz | 85.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18136-v30.xml emd-18136-v30.xml emd-18136.xml emd-18136.xml | 27.3 KB 27.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18136_fsc.xml emd_18136_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_18136.png emd_18136.png | 110.2 KB | ||
| Masks |  emd_18136_msk_1.map emd_18136_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18136.cif.gz emd-18136.cif.gz | 6.9 KB | ||
| Others |  emd_18136_additional_1.map.gz emd_18136_additional_1.map.gz emd_18136_additional_2.map.gz emd_18136_additional_2.map.gz emd_18136_half_map_1.map.gz emd_18136_half_map_1.map.gz emd_18136_half_map_2.map.gz emd_18136_half_map_2.map.gz | 69.6 MB 2.5 MB 70 MB 70 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18136 http://ftp.pdbj.org/pub/emdb/structures/EMD-18136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18136 | HTTPS FTP |
-Validation report
| Summary document |  emd_18136_validation.pdf.gz emd_18136_validation.pdf.gz | 905.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18136_full_validation.pdf.gz emd_18136_full_validation.pdf.gz | 905.4 KB | Display | |
| Data in XML |  emd_18136_validation.xml.gz emd_18136_validation.xml.gz | 17.2 KB | Display | |
| Data in CIF |  emd_18136_validation.cif.gz emd_18136_validation.cif.gz | 22.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18136 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18136 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18136 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18136 | HTTPS FTP |
-Related structure data
| Related structure data |  8q3wMC  8q4dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18136.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18136.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
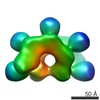
| Annotation | Sharpened Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18136_msk_1.map emd_18136_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened Map
| File | emd_18136_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map used for modelling of amino-terminal domains after...
| File | emd_18136_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map used for modelling of amino-terminal domains after focused 3D classification | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18136_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18136_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : IstB transposition regulator AAA+ ATPase bound to target DNA
| Entire | Name: IstB transposition regulator AAA+ ATPase bound to target DNA |
|---|---|
| Components |
|
-Supramolecule #1: IstB transposition regulator AAA+ ATPase bound to target DNA
| Supramolecule | Name: IstB transposition regulator AAA+ ATPase bound to target DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Complex of IstB transposition regulator AAA+ ATPase bound to target DNA |
|---|---|
| Molecular weight | Theoretical: 337 KDa |
-Supramolecule #2: Insertion sequence IS5376 putative ATP-binding protein
| Supramolecule | Name: Insertion sequence IS5376 putative ATP-binding protein type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
-Supramolecule #3: DNA (48-MER) Target DNA FW and Rv
| Supramolecule | Name: DNA (48-MER) Target DNA FW and Rv / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) / Synthetically produced: Yes Geobacillus stearothermophilus (bacteria) / Synthetically produced: Yes |
-Macromolecule #1: Insertion sequence IS5376 putative ATP-binding protein
| Macromolecule | Name: Insertion sequence IS5376 putative ATP-binding protein type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 29.602275 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPNMKERIHE YCHRLHLPVM AERWSAMAEY ASTHNISYSE FLFRLLEAEI VEKQARSIQT LIKLSKLPYR KTIDTFDFTA QPSVDERRI RELLTLSFID RKENILFLGP PGIGKTHLAI SIGMEAIARG YKTYFITAHD LVNQLRRADQ EGKLEKKLRV F VKPTVLII ...String: GPNMKERIHE YCHRLHLPVM AERWSAMAEY ASTHNISYSE FLFRLLEAEI VEKQARSIQT LIKLSKLPYR KTIDTFDFTA QPSVDERRI RELLTLSFID RKENILFLGP PGIGKTHLAI SIGMEAIARG YKTYFITAHD LVNQLRRADQ EGKLEKKLRV F VKPTVLII DEMGYLKLDP NSAHYLFQVI ARRYEHAPII LTSNKSFGEW GEIVGDSVLA TAMLDRLLHH SIIFNLKGES YR LREKRLQ EEKQKDQ UniProtKB: Insertion sequence IS5376 putative ATP-binding protein |
-Macromolecule #2: DNA (48-MER) Target DNA FW
| Macromolecule | Name: DNA (48-MER) Target DNA FW / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 18.401746 KDa |
| Sequence | String: (DT)(DG)(DC)(DT)(DT)(DG)(DC)(DG)(DA)(DT) (DG)(DA)(DT)(DC)(DC)(DG)(DA)(DC)(DG)(DT) (DG)(DT)(DT)(DA)(DG)(DC)(DC)(DA)(DC) (DG)(DC)(DT)(DG)(DA)(DC)(DT)(DA)(DG)(DT) (DT) (DA)(DT)(DG)(DC)(DC)(DA) ...String: (DT)(DG)(DC)(DT)(DT)(DG)(DC)(DG)(DA)(DT) (DG)(DA)(DT)(DC)(DC)(DG)(DA)(DC)(DG)(DT) (DG)(DT)(DT)(DA)(DG)(DC)(DC)(DA)(DC) (DG)(DC)(DT)(DG)(DA)(DC)(DT)(DA)(DG)(DT) (DT) (DA)(DT)(DG)(DC)(DC)(DA)(DT)(DG) (DC)(DC)(DT)(DC)(DC)(DC)(DT)(DT)(DC)(DA) (DG)(DG) |
-Macromolecule #3: DNA (48-MER) Traget DNA Rv
| Macromolecule | Name: DNA (48-MER) Traget DNA Rv / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 18.584918 KDa |
| Sequence | String: (DC)(DC)(DT)(DG)(DA)(DA)(DG)(DG)(DG)(DA) (DG)(DG)(DC)(DA)(DT)(DG)(DG)(DC)(DA)(DT) (DA)(DA)(DC)(DT)(DA)(DG)(DT)(DC)(DA) (DG)(DC)(DG)(DT)(DG)(DG)(DC)(DT)(DA)(DA) (DC) (DA)(DC)(DG)(DT)(DC)(DG) ...String: (DC)(DC)(DT)(DG)(DA)(DA)(DG)(DG)(DG)(DA) (DG)(DG)(DC)(DA)(DT)(DG)(DG)(DC)(DA)(DT) (DA)(DA)(DC)(DT)(DA)(DG)(DT)(DC)(DA) (DG)(DC)(DG)(DT)(DG)(DG)(DC)(DT)(DA)(DA) (DC) (DA)(DC)(DG)(DT)(DC)(DG)(DG)(DA) (DT)(DC)(DA)(DT)(DC)(DG)(DC)(DA)(DA)(DG) (DC)(DA) |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 10 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 10 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 5.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 95.21 |
|---|---|
| Output model |  PDB-8q3w: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)