+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1810 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phage T7 empty head. | |||||||||
 Map data Map data | Bacteriophage T7 empty head | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacteriophage / virus / capsid maturation / cryoelectron microscopy / morphogenetic intermediate. | |||||||||
| Function / homology | Capsid Gp10A/Gp10B / : / Major capsid protein / viral capsid / viral translational frameshifting / identical protein binding / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.8 Å | |||||||||
 Authors Authors | Ionel A / Velazquez-Muriel JA / Luque D / Cuervo A / Caston JR / Valpuesta JM / Martin-Benito J / Carrascosa JL | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2011 Journal: J Biol Chem / Year: 2011Title: Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. Authors: Alina Ionel / Javier A Velázquez-Muriel / Daniel Luque / Ana Cuervo / José R Castón / José M Valpuesta / Jaime Martín-Benito / José L Carrascosa /  Abstract: Maturation of dsDNA bacteriophages involves assembling the virus prohead from a limited set of structural components followed by rearrangements required for the stability that is necessary for ...Maturation of dsDNA bacteriophages involves assembling the virus prohead from a limited set of structural components followed by rearrangements required for the stability that is necessary for infecting a host under challenging environmental conditions. Here, we determine the mature capsid structure of T7 at 1 nm resolution by cryo-electron microscopy and compare it with the prohead to reveal the molecular basis of T7 shell maturation. The mature capsid presents an expanded and thinner shell, with a drastic rearrangement of the major protein monomers that increases in their interacting surfaces, in turn resulting in a new bonding lattice. The rearrangements include tilting, in-plane rotation, and radial expansion of the subunits, as well as a relative bending of the A- and P-domains of each subunit. The unique features of this shell transformation, which does not employ the accessory proteins, inserted domains, or molecular interactions observed in other phages, suggest a simple capsid assembling strategy that may have appeared early in the evolution of these viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1810.map.gz emd_1810.map.gz | 597.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1810-v30.xml emd-1810-v30.xml emd-1810.xml emd-1810.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  1810_T7matureHead.jpg 1810_T7matureHead.jpg | 456.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1810 http://ftp.pdbj.org/pub/emdb/structures/EMD-1810 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1810 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1810 | HTTPS FTP |
-Related structure data
| Related structure data |  2xvrMC  3izgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1810.map.gz / Format: CCP4 / Size: 626.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1810.map.gz / Format: CCP4 / Size: 626.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacteriophage T7 empty head | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
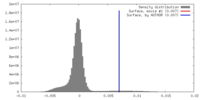
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Phage T7 empty head
| Entire | Name: Phage T7 empty head |
|---|---|
| Components |
|
-Supramolecule #1000: Phage T7 empty head
| Supramolecule | Name: Phage T7 empty head / type: sample / ID: 1000 / Oligomeric state: Icosahedral / Number unique components: 1 |
|---|
-Supramolecule #1: Enterobacteria phage T7
| Supramolecule | Name: Enterobacteria phage T7 / type: virus / ID: 1 / Name.synonym: Bacteriophage T7 / NCBI-ID: 10760 / Sci species name: Enterobacteria phage T7 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes / Syn species name: Bacteriophage T7 |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 600 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 / Details: 50 mM Tris-HCl, pH 7.8, 10 mM MgCl2, 0.1 M NaCl |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 10.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: xmipp / Number images used: 5100 |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)