[English] 日本語
 Yorodumi
Yorodumi- EMDB-17553: Cryo-EM structure of cortactin-stabilized Arp2/3 complex nucleate... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of cortactin-stabilized Arp2/3 complex nucleated actin branches-Daughter filament consensus map | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / CONTRACTILE PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
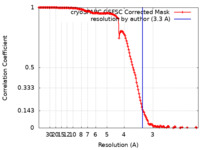
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Liu T / Moores CA | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Cortactin stabilizes actin branches by bridging activated Arp2/3 to its nucleated actin filament. Authors: Tianyang Liu / Luyan Cao / Miroslav Mladenov / Antoine Jegou / Michael Way / Carolyn A Moores /   Abstract: Regulation of the assembly and turnover of branched actin filament networks nucleated by the Arp2/3 complex is essential during many cellular processes, including cell migration and membrane ...Regulation of the assembly and turnover of branched actin filament networks nucleated by the Arp2/3 complex is essential during many cellular processes, including cell migration and membrane trafficking. Cortactin is important for actin branch stabilization, but the mechanism by which this occurs is unclear. Given this, we determined the structure of vertebrate cortactin-stabilized Arp2/3 actin branches using cryogenic electron microscopy. We find that cortactin interacts with the new daughter filament nucleated by the Arp2/3 complex at the branch site, rather than the initial mother actin filament. Cortactin preferentially binds activated Arp3. It also stabilizes the F-actin-like interface of activated Arp3 with the first actin subunit of the new filament, and its central repeats extend along successive daughter-filament subunits. The preference of cortactin for activated Arp3 explains its retention at the actin branch and accounts for its synergy with other nucleation-promoting factors in regulating branched actin network dynamics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17553.map.gz emd_17553.map.gz | 163.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17553-v30.xml emd-17553-v30.xml emd-17553.xml emd-17553.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17553_fsc.xml emd_17553_fsc.xml | 16.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17553.png emd_17553.png | 69.9 KB | ||
| Filedesc metadata |  emd-17553.cif.gz emd-17553.cif.gz | 4.4 KB | ||
| Others |  emd_17553_half_map_1.map.gz emd_17553_half_map_1.map.gz emd_17553_half_map_2.map.gz emd_17553_half_map_2.map.gz | 301.2 MB 301.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17553 http://ftp.pdbj.org/pub/emdb/structures/EMD-17553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17553 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17553.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17553.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||
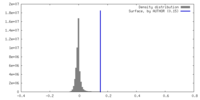
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_17553_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
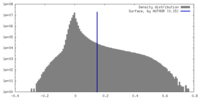
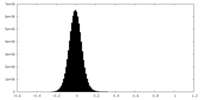
| Density Histograms |
-Half map: #2
| File | emd_17553_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
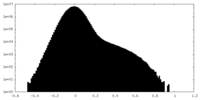
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of cortactin-stabilized Arp2/3 complex nucleate...
| Entire | Name: Cryo-EM structure of cortactin-stabilized Arp2/3 complex nucleated actin branches |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of cortactin-stabilized Arp2/3 complex nucleate...
| Supramolecule | Name: Cryo-EM structure of cortactin-stabilized Arp2/3 complex nucleated actin branches type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#10 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 20 mM HEPES pH 7.5, 50mM KCl, 1mM EGTA, 1mM MgCl2, 0.2 mM ATP and 1 mM DTT |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 295.15 K / Instrument: LEICA EM GP / Details: Back blotting. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 49.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Protocol: OTHER |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)