[English] 日本語
 Yorodumi
Yorodumi- EMDB-17421: CryoEM structure of a GroEL7-GroES7 cage with encapsulated ordere... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of a GroEL7-GroES7 cage with encapsulated ordered substrate MetK in the presence of ADP-BeFx | |||||||||
 Map data Map data | GroEL7-GroES7 cage with encapsulated ordered substrate MetK in presence of ADP-BeFx | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Chaperonin / Folding cage / proteostasis / heat shock / ATPase / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmethionine adenosyltransferase / methionine adenosyltransferase activity / S-adenosylmethionine biosynthetic process / S-adenosylmethionine cycle / GroEL-GroES complex / chaperonin ATPase / virion assembly / : / potassium ion binding / isomerase activity ...methionine adenosyltransferase / methionine adenosyltransferase activity / S-adenosylmethionine biosynthetic process / S-adenosylmethionine cycle / GroEL-GroES complex / chaperonin ATPase / virion assembly / : / potassium ion binding / isomerase activity / one-carbon metabolic process / protein folding chaperone / ATP-dependent protein folding chaperone / response to radiation / unfolded protein binding / protein-folding chaperone binding / protein folding / response to heat / protein refolding / magnesium ion binding / ATP hydrolysis activity / ATP binding / metal ion binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
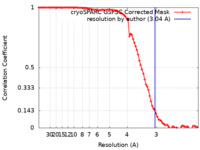
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Wagner J / Beck F / Bracher A / Caravajal AI / Wan W / Bohn S / Koerner R / Baumeister W / Fernandez-Busnadiego R / Hartl FU | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Visualizing chaperonin function in situ by cryo-electron tomography. Authors: Jonathan Wagner / Alonso I Carvajal / Andreas Bracher / Florian Beck / William Wan / Stefan Bohn / Roman Körner / Wolfgang Baumeister / Ruben Fernandez-Busnadiego / F Ulrich Hartl /   Abstract: Chaperonins are large barrel-shaped complexes that mediate ATP-dependent protein folding. The bacterial chaperonin GroEL forms juxtaposed rings that bind unfolded protein and the lid-shaped cofactor ...Chaperonins are large barrel-shaped complexes that mediate ATP-dependent protein folding. The bacterial chaperonin GroEL forms juxtaposed rings that bind unfolded protein and the lid-shaped cofactor GroES at their apertures. In vitro analyses of the chaperonin reaction have shown that substrate protein folds, unimpaired by aggregation, while transiently encapsulated in the GroEL central cavity by GroES. To determine the functional stoichiometry of GroEL, GroES and client protein in situ, here we visualized chaperonin complexes in their natural cellular environment using cryo-electron tomography. We find that, under various growth conditions, around 55-70% of GroEL binds GroES asymmetrically on one ring, with the remainder populating symmetrical complexes. Bound substrate protein is detected on the free ring of the asymmetrical complex, defining the substrate acceptor state. In situ analysis of GroEL-GroES chambers, validated by high-resolution structures obtained in vitro, showed the presence of encapsulated substrate protein in a folded state before release into the cytosol. Based on a comprehensive quantification and conformational analysis of chaperonin complexes, we propose a GroEL-GroES reaction cycle that consists of linked asymmetrical and symmetrical subreactions mediating protein folding. Our findings illuminate the native conformational and functional chaperonin cycle directly within cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17421.map.gz emd_17421.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17421-v30.xml emd-17421-v30.xml emd-17421.xml emd-17421.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17421_fsc.xml emd_17421_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17421.png emd_17421.png | 101.9 KB | ||
| Filedesc metadata |  emd-17421.cif.gz emd-17421.cif.gz | 7.1 KB | ||
| Others |  emd_17421_half_map_1.map.gz emd_17421_half_map_1.map.gz emd_17421_half_map_2.map.gz emd_17421_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17421 http://ftp.pdbj.org/pub/emdb/structures/EMD-17421 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17421 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17421 | HTTPS FTP |
-Related structure data
| Related structure data |  8p4oMC  8p4mC  8p4nC  8p4pC  8p4rC  8qxsC  8qxtC  8qxuC  8qxvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17421.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17421.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GroEL7-GroES7 cage with encapsulated ordered substrate MetK in presence of ADP-BeFx | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_17421_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17421_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GroEL14-GroES14-MetK2
| Entire | Name: GroEL14-GroES14-MetK2 |
|---|---|
| Components |
|
-Supramolecule #1: GroEL14-GroES14-MetK2
| Supramolecule | Name: GroEL14-GroES14-MetK2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / Details: with bound ADP-BeF3 Mg2+ K+ |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.03 MDa |
-Macromolecule #1: Chaperonin GroEL
| Macromolecule | Name: Chaperonin GroEL / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO / EC number: chaperonin ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.260504 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AAKDVKFGND ARVKMLRGVN VLADAVKVTL GPKGRNVVLD KSFGAPTITK DGVSVAREIE LEDKFENMGA QMVKEVASKA NDAAGDGTT TATVLAQAII TEGLKAVAAG MNPMDLKRGI DKAVTAAVEE LKALSVPCSD SKAIAQVGTI SANSDETVGK L IAEAMDKV ...String: AAKDVKFGND ARVKMLRGVN VLADAVKVTL GPKGRNVVLD KSFGAPTITK DGVSVAREIE LEDKFENMGA QMVKEVASKA NDAAGDGTT TATVLAQAII TEGLKAVAAG MNPMDLKRGI DKAVTAAVEE LKALSVPCSD SKAIAQVGTI SANSDETVGK L IAEAMDKV GKEGVITVED GTGLQDELDV VEGMQFDRGY LSPYFINKPE TGAVELESPF ILLADKKISN IREMLPVLEA VA KAGKPLL IIAEDVEGEA LATLVVNTMR GIVKVAAVKA PGFGDRRKAM LQDIATLTGG TVISEEIGME LEKATLEDLG QAK RVVINK DTTTIIDGVG EEAAIQGRVA QIRQQIEEAT SDYDREKLQE RVAKLAGGVA VIKVGAATEV EMKEKKARVE DALH ATRAA VEEGVVAGGG VALIRVASKL ADLRGQNEDQ NVGIKVALRA MEAPLRQIVL NCGEEPSVVA NTVKGGDGNY GYNAA TEEY GNMIDMGILD PTKVTRSALQ YAASVAGLMI TTECMVTDLP KNDAADLGAA GGMGGMGGMG GMM UniProtKB: Chaperonin GroEL |
-Macromolecule #2: Co-chaperonin GroES
| Macromolecule | Name: Co-chaperonin GroES / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.400938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNIRPLHDRV IVKRKEVETK SAGGIVLTGS AAAKSTRGEV LAVGNGRILE NGEVKPLDVK VGDIVIFNDG YGVKSEKIDN EEVLIMSES DILAIVEA UniProtKB: Co-chaperonin GroES |
-Macromolecule #3: S-adenosylmethionine synthase
| Macromolecule | Name: S-adenosylmethionine synthase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: methionine adenosyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.998508 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKHLFTSES VSEGHPDKIA DQISDAVLDA ILEQDPKARV ACETYVKTGM VLVGGEITTS AWVDIEEITR NTVREIGYVH SDMGFDANS CAVLSAIGKQ SPDINQGVDR ADPLEQGAGD QGLMFGYATN ETDVLMPAPI TYAHRLVQRQ AEVRKNGTLP W LRPDAKSQ ...String: MAKHLFTSES VSEGHPDKIA DQISDAVLDA ILEQDPKARV ACETYVKTGM VLVGGEITTS AWVDIEEITR NTVREIGYVH SDMGFDANS CAVLSAIGKQ SPDINQGVDR ADPLEQGAGD QGLMFGYATN ETDVLMPAPI TYAHRLVQRQ AEVRKNGTLP W LRPDAKSQ VTFQYDDGKI VGIDAVVLST QHSEEIDQKS LQEAVMEEII KPILPAEWLT SATKFFINPT GRFVIGGPMG DC GLTGRKI IVDTYGGMAR HGGGAFSGKD PSKVDRSAAY AARYVAKNIV AAGLADRCEI QVSYAIGVAE PTSIMVETFG TEK VPSEQL TLLVREFFDL RPYGLIQMLD LLHPIYKETA AYGHFGREHF PWEKTDKAQL LRDAAGLK UniProtKB: S-adenosylmethionine synthase |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 7 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 7 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: BERYLLIUM TRIFLUORIDE ION
| Macromolecule | Name: BERYLLIUM TRIFLUORIDE ION / type: ligand / ID: 6 / Number of copies: 7 / Formula: BEF |
|---|---|
| Molecular weight | Theoretical: 66.007 Da |
| Chemical component information |  ChemComp-BEF: |
-Macromolecule #7: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 7 / Number of copies: 7 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 21 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10.3 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 5.9 mM n-octyl-beta-D-glucopyranoside were added before vitrification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL | ||||||
| Output model |  PDB-8p4o: |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)