[English] 日本語
 Yorodumi
Yorodumi- EMDB-17262: Cryo-EM structure of ATP8B1-CDC50A in E2-Pi conformation with occ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATP8B1-CDC50A in E2-Pi conformation with occluded PS | |||||||||||||||||||||||||||
 Map data Map data | sharpened map by PHENIX autosharpen B factor applied 65 | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | lipid transporter autoinhibition P-type ATPase P4-ATPase CDC50A / MEMBRANE PROTEIN | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationvestibulocochlear nerve formation / regulation of plasma membrane organization / regulation of microvillus assembly / phosphatidylcholine flippase activity / positive regulation of phospholipid translocation / aminophospholipid flippase activity / aminophospholipid transport / phosphatidylserine flippase activity / protein localization to endosome / phospholipid-translocating ATPase complex ...vestibulocochlear nerve formation / regulation of plasma membrane organization / regulation of microvillus assembly / phosphatidylcholine flippase activity / positive regulation of phospholipid translocation / aminophospholipid flippase activity / aminophospholipid transport / phosphatidylserine flippase activity / protein localization to endosome / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / positive regulation of protein exit from endoplasmic reticulum / ATPase-coupled intramembrane lipid transporter activity / phosphatidylcholine floppase activity / xenobiotic transmembrane transport / stereocilium / cardiolipin binding / bile acid metabolic process / P-type phospholipid transporter / inner ear receptor cell development / apical protein localization / phospholipid translocation / azurophil granule membrane / bile acid and bile salt transport / transport vesicle membrane / Golgi organization / Ion transport by P-type ATPases / specific granule membrane / regulation of chloride transport / trans-Golgi network / sensory perception of sound / positive regulation of neuron projection development / late endosome membrane / early endosome membrane / monoatomic ion transmembrane transport / apical plasma membrane / nuclear body / negative regulation of DNA-templated transcription / Neutrophil degranulation / structural molecule activity / magnesium ion binding / endoplasmic reticulum / Golgi apparatus / ATP hydrolysis activity / nucleoplasm / ATP binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
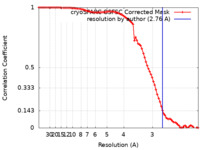
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||||||||||||||||||||
 Authors Authors | Dieudonne T / Kummerer F / Juknaviciute Laursen M / Stock C / Kock Flygaard R / Khalid S / Lenoir G / Lyons JA / Lindorff-Larsen K / Nissen P | |||||||||||||||||||||||||||
| Funding support | European Union,  Denmark, Denmark,  France, France,  United Kingdom, 8 items United Kingdom, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Activation and substrate specificity of the human P4-ATPase ATP8B1. Authors: Thibaud Dieudonné / Felix Kümmerer / Michelle Juknaviciute Laursen / Charlott Stock / Rasmus Kock Flygaard / Syma Khalid / Guillaume Lenoir / Joseph A Lyons / Kresten Lindorff-Larsen / Poul Nissen /    Abstract: Asymmetric distribution of phospholipids in eukaryotic membranes is essential for cell integrity, signaling pathways, and vesicular trafficking. P4-ATPases, also known as flippases, participate in ...Asymmetric distribution of phospholipids in eukaryotic membranes is essential for cell integrity, signaling pathways, and vesicular trafficking. P4-ATPases, also known as flippases, participate in creating and maintaining this asymmetry through active transport of phospholipids from the exoplasmic to the cytosolic leaflet. Here, we present a total of nine cryo-electron microscopy structures of the human flippase ATP8B1-CDC50A complex at 2.4 to 3.1 Å overall resolution, along with functional and computational studies, addressing the autophosphorylation steps from ATP, substrate recognition and occlusion, as well as a phosphoinositide binding site. We find that the P4-ATPase transport site is occupied by water upon phosphorylation from ATP. Additionally, we identify two different autoinhibited states, a closed and an outward-open conformation. Furthermore, we identify and characterize the PI(3,4,5)P binding site of ATP8B1 in an electropositive pocket between transmembrane segments 5, 7, 8, and 10. Our study also highlights the structural basis of a broad lipid specificity of ATP8B1 and adds phosphatidylinositol as a transport substrate for ATP8B1. We report a critical role of the sn-2 ester bond of glycerophospholipids in substrate recognition by ATP8B1 through conserved S403. These findings provide fundamental insights into ATP8B1 catalytic cycle and regulation, and substrate recognition in P4-ATPases. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17262.map.gz emd_17262.map.gz | 56.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17262-v30.xml emd-17262-v30.xml emd-17262.xml emd-17262.xml | 23.9 KB 23.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17262_fsc.xml emd_17262_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17262.png emd_17262.png | 134.2 KB | ||
| Filedesc metadata |  emd-17262.cif.gz emd-17262.cif.gz | 7.6 KB | ||
| Others |  emd_17262_additional_1.map.gz emd_17262_additional_1.map.gz emd_17262_half_map_1.map.gz emd_17262_half_map_1.map.gz emd_17262_half_map_2.map.gz emd_17262_half_map_2.map.gz | 32 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17262 http://ftp.pdbj.org/pub/emdb/structures/EMD-17262 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17262 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17262 | HTTPS FTP |
-Related structure data
| Related structure data |  8oxaMC  8ox4C  8ox5C  8ox6C  8ox7C  8ox8C  8ox9C  8oxbC  8oxcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17262.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17262.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map by PHENIX autosharpen B factor applied 65 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05138 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: raw map before sharpening
| File | emd_17262_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | raw map before sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
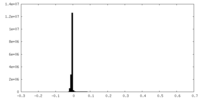
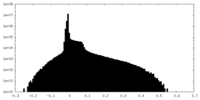
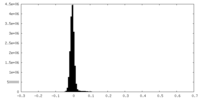
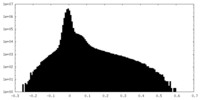
| Density Histograms |
-Half map: half map A
| File | emd_17262_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_17262_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
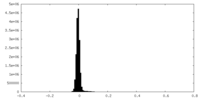
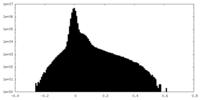
| Density Histograms |
- Sample components
Sample components
-Entire : ATP8B1-CDC50A complex
| Entire | Name: ATP8B1-CDC50A complex |
|---|---|
| Components |
|
-Supramolecule #1: ATP8B1-CDC50A complex
| Supramolecule | Name: ATP8B1-CDC50A complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 185 KDa |
-Macromolecule #1: Phospholipid-transporting ATPase IC
| Macromolecule | Name: Phospholipid-transporting ATPase IC / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: P-type phospholipid transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 136.283469 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTERDSETT FDEDSQPNDE VVPYSDDETE DELDDQGSAV EPEQNRVNRE AEENREPFRK ECTWQVKAND RKYHEQPHFM NTKFLCIKE SKYANNAIKT YKYNAFTFIP MNLFEQFKRA ANLYFLALLI LQAVPQISTL AWYTTLVPLL VVLGVTAIKD L VDDVARHK ...String: MSTERDSETT FDEDSQPNDE VVPYSDDETE DELDDQGSAV EPEQNRVNRE AEENREPFRK ECTWQVKAND RKYHEQPHFM NTKFLCIKE SKYANNAIKT YKYNAFTFIP MNLFEQFKRA ANLYFLALLI LQAVPQISTL AWYTTLVPLL VVLGVTAIKD L VDDVARHK MDKEINNRTC EVIKDGRFKV AKWKEIQVGD VIRLKKNDFV PADILLLSSS EPNSLCYVET AELDGETNLK FK MSLEITD QYLQREDTLA TFDGFIECEE PNNRLDKFTG TLFWRNTSFP LDADKILLRG CVIRNTDFCH GLVIFAGADT KIM KNSGKT RFKRTKIDYL MNYMVYTIFV VLILLSAGLA IGHAYWEAQV GNSSWYLYDG EDDTPSYRGF LIFWGYIIVL NTMV PISLY VSVEVIRLGQ SHFINWDLQM YYAEKDTPAK ARTTTLNEQL GQIHYIFSDK TGTLTQNIMT FKKCCINGQI YGDHR DASQ HNHNKIEQVD FSWNTYADGK LAFYDHYLIE QIQSGKEPEV RQFFFLLAVC HTVMVDRTDG QLNYQAASPD EGALVN AAR NFGFAFLART QNTITISELG TERTYNVLAI LDFNSDRKRM SIIVRTPEGN IKLYCKGADT VIYERLHRMN PTKQETQ DA LDIFANETLR TLCLCYKEIE EKEFTEWNKK FMAASVASTN RDEALDKVYE EIEKDLILLG ATAIEDKLQD GVPETISK L AKADIKIWVL TGDKKETAEN IGFACELLTE DTTICYGEDI NSLLHARMEN QRNRGGVYAK FAPPVQESFF PPGGNRALI ITGSWLNEIL LEKKTKRNKI LKLKFPRTEE ERRMRTQSKR RLEAKKEQRQ KNFVDLACEC SAVICCRVTP KQKAMVVDLV KRYKKAITL AIGDGANDVN MIKTAHIGVG ISGQEGMQAV MSSDYSFAQF RYLQRLLLVH GRWSYIRMCK FLRYFFYKNF A FTLVHFWY SFFNGYSAQT AYEDWFITLY NVLYTSLPVL LMGLLDQDVS DKLSLRFPGL YIVGQRDLLF NYKRFFVSLL HG VLTSMIL FFIPLGAYLQ TVGQDGEAPS DYQSFAVTIA SALVITVNFQ IGLDTSYWTF VNAFSIFGSI ALYFGIMFDF HSA GIHVLF PSAFQFTGTA SNALRQPYIW LTIILTVAVC LLPVVAIRFL SMTIWPSESD KIQKHRKRL UniProtKB: Phospholipid-transporting ATPase IC |
-Macromolecule #2: Cell cycle control protein 50A
| Macromolecule | Name: Cell cycle control protein 50A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.085984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLGGMAMNYN AKDEVDGGPP CAPGGTAKTR RPDNTAFKQQ RLPAWQPILT AGTVLPIFFI IGLIFIPIGI GIFVTSNNIR EIEIDYTGT EPSSPCNKCL SPDVTPCFCT INFTLEKSFE GNVFMYYGLS NFYQNHRRYV KSRDDSQLNG DSSALLNPSK E CEPYRRNE ...String: MLGGMAMNYN AKDEVDGGPP CAPGGTAKTR RPDNTAFKQQ RLPAWQPILT AGTVLPIFFI IGLIFIPIGI GIFVTSNNIR EIEIDYTGT EPSSPCNKCL SPDVTPCFCT INFTLEKSFE GNVFMYYGLS NFYQNHRRYV KSRDDSQLNG DSSALLNPSK E CEPYRRNE DKPIAPCGAI ANSMFNDTLE LFLIGNDSYP IPIALKKKGI AWWTDKNVKF RNPPGGDNLE ERFKGTTKPV NW LKPVYML DSDPDNNGFI NEDFIVWMRT AALPTFRKLY RLIERKSDLH PTLPAGRYSL NVTYNYPVHY FDGRKRMILS TIS WMGGKN PFLGIAYIAV GSISFLLGVV LLVINHKYRN SSNTADITI UniProtKB: Cell cycle control protein 50A |
-Macromolecule #4: oxido(dioxo)vanadium
| Macromolecule | Name: oxido(dioxo)vanadium / type: ligand / ID: 4 / Number of copies: 1 / Formula: VN4 |
|---|---|
| Molecular weight | Theoretical: 98.94 Da |
| Chemical component information |  ChemComp-VN4: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-...
| Macromolecule | Name: (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: D39 |
|---|---|
| Molecular weight | Theoretical: 762.006 Da |
| Chemical component information |  ChemComp-D39: |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 11 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | 50 mM MOPS-Tris pH 7, 100 mM KCl, 1 mM DTT, 5 mM MgCl2 supplemented with 0.03 mg.mL-1 LMNG, 0.003 mg.mL-1 CHS, PI(4,5)P2 0.0015 mg.mL-1 and 1 mM Na3VO4 and 0.003 mg.mL-1 POPS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)