[English] 日本語
 Yorodumi
Yorodumi- EMDB-16699: Subtomogram average of HIV-1 CA hexamer from capsid-like particle... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of HIV-1 CA hexamer from capsid-like particles assembled with inositol hexakisphosphate | |||||||||||||||||||||||||||
 Map data Map data | Subtomogram average of HIV-1 CA protein hexamer from capsid-like particles assembled with inositol hexakisphosphate | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 3.9 Å | |||||||||||||||||||||||||||
 Authors Authors | Tan A / Briggs JAG / Dick RA | |||||||||||||||||||||||||||
| Funding support |  United States, United States,  United Kingdom, United Kingdom,  Germany, 8 items Germany, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM. Authors: Carolyn M Highland / Aaron Tan / Clifton L Ricaña / John A G Briggs / Robert A Dick /    Abstract: The HIV-1 capsid houses the viral genome and interacts extensively with host cell proteins throughout the viral life cycle. It is composed of capsid protein (CA), which assembles into a conical ...The HIV-1 capsid houses the viral genome and interacts extensively with host cell proteins throughout the viral life cycle. It is composed of capsid protein (CA), which assembles into a conical fullerene lattice composed of roughly 200 CA hexamers and 12 CA pentamers. Previous structural analyses of individual CA hexamers and pentamers have provided valuable insight into capsid structure and function, but detailed structural information about these assemblies in the broader context of the capsid lattice is lacking. In this study, we combined cryoelectron tomography and single particle analysis (SPA) cryoelectron microscopy to determine structures of continuous regions of the capsid lattice containing both hexamers and pentamers. We also developed a method of liposome scaffold-based in vitro lattice assembly ("lattice templating") that enabled us to directly study the lattice under a wider range of conditions than has previously been possible. Using this approach, we identified a critical role for inositol hexakisphosphate in pentamer formation and determined the structure of the CA lattice bound to the capsid-targeting antiretroviral drug GS-6207 (lenacapavir). Our work reveals key structural details of the mature HIV-1 CA lattice and establishes the combination of lattice templating and SPA as a robust strategy for studying retroviral capsid structure and capsid interactions with host proteins and antiviral compounds. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16699.map.gz emd_16699.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16699-v30.xml emd-16699-v30.xml emd-16699.xml emd-16699.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16699.png emd_16699.png | 144.8 KB | ||
| Others |  emd_16699_half_map_1.map.gz emd_16699_half_map_1.map.gz emd_16699_half_map_2.map.gz emd_16699_half_map_2.map.gz | 25.1 MB 25.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16699 http://ftp.pdbj.org/pub/emdb/structures/EMD-16699 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16699 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16699 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16699.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16699.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of HIV-1 CA protein hexamer from capsid-like particles assembled with inositol hexakisphosphate | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.379 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Subtomogram average of HIV-1 CA protein hexamer from...
| File | emd_16699_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of HIV-1 CA protein hexamer from capsid-like particles assembled with inositol hexakisphosphate, half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
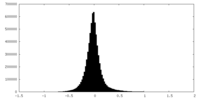
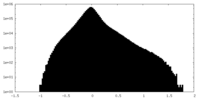
| Density Histograms |
-Half map: Subtomogram average of HIV-1 CA protein hexamer from...
| File | emd_16699_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of HIV-1 CA protein hexamer from capsid-like particles assembled with inositol hexakisphosphate, half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
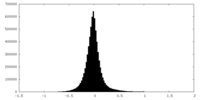
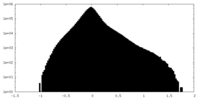
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 CA protein capsid-like particles assembled in the presence ...
| Entire | Name: HIV-1 CA protein capsid-like particles assembled in the presence of inositol hexakisphosphate |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 CA protein capsid-like particles assembled in the presence ...
| Supramolecule | Name: HIV-1 CA protein capsid-like particles assembled in the presence of inositol hexakisphosphate type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.2 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR Details: The grid was glow discharged for 30 seconds at a current of 20 mA. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: subTOM / Number subtomograms used: 89951 |
|---|---|
| Extraction | Number tomograms: 66 / Number images used: 570742 / Software - Name:  MATLAB MATLABDetails: HIV-1 CA protein capsid-like particles (CLPs) were segmented using the Ilastik software package. The vertex coordinates of the segmented volumes were used to define an oversampled grid of ...Details: HIV-1 CA protein capsid-like particles (CLPs) were segmented using the Ilastik software package. The vertex coordinates of the segmented volumes were used to define an oversampled grid of points normal to the CLP surface in MATLAB. |
| Final angle assignment | Type: OTHER / Software - Name: subTOM |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)