+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM Map of the latTGF-beta LHG-10 Fab complex | |||||||||
 マップデータ マップデータ | Sharpened map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Fab-complex / GARP / lat-TGF-beta / IMMUNE SYSTEM | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報establishment of protein localization to extracellular region / cellular response to acetaldehyde / frontal suture morphogenesis / Influenza Virus Induced Apoptosis / adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains / positive regulation of microglia differentiation / regulation of interleukin-23 production / branch elongation involved in mammary gland duct branching / positive regulation of primary miRNA processing / columnar/cuboidal epithelial cell maturation ...establishment of protein localization to extracellular region / cellular response to acetaldehyde / frontal suture morphogenesis / Influenza Virus Induced Apoptosis / adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains / positive regulation of microglia differentiation / regulation of interleukin-23 production / branch elongation involved in mammary gland duct branching / positive regulation of primary miRNA processing / columnar/cuboidal epithelial cell maturation / negative regulation of skeletal muscle tissue development / macrophage derived foam cell differentiation / response to laminar fluid shear stress / embryonic liver development / regulation of enamel mineralization / regulation of branching involved in mammary gland duct morphogenesis / regulation of cartilage development / TGFBR2 MSI Frameshift Mutants in Cancer / regulation of striated muscle tissue development / regulation of blood vessel remodeling / regulation of protein import into nucleus / regulatory T cell differentiation / tolerance induction to self antigen / extracellular matrix assembly / negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / negative regulation of hyaluronan biosynthetic process / type III transforming growth factor beta receptor binding / myofibroblast differentiation / positive regulation of cardiac muscle cell differentiation / positive regulation of odontogenesis / connective tissue replacement involved in inflammatory response wound healing / Langerhans cell differentiation / TGFBR2 Kinase Domain Mutants in Cancer / positive regulation of smooth muscle cell differentiation / positive regulation of exit from mitosis / secondary palate development / negative regulation of macrophage cytokine production / odontoblast differentiation / SMAD2/3 Phosphorylation Motif Mutants in Cancer / TGFBR1 KD Mutants in Cancer / positive regulation of isotype switching to IgA isotypes / positive regulation of mesenchymal stem cell proliferation / positive regulation of receptor signaling pathway via STAT / membrane protein intracellular domain proteolysis / positive regulation of extracellular matrix assembly / retina vasculature development in camera-type eye / heart valve morphogenesis / bronchiole development / mammary gland branching involved in thelarche / hyaluronan catabolic process / TGFBR3 regulates TGF-beta signaling / positive regulation of vasculature development / lens fiber cell differentiation / ATP biosynthetic process / negative regulation of extracellular matrix disassembly / type II transforming growth factor beta receptor binding / receptor catabolic process / positive regulation of branching involved in ureteric bud morphogenesis / TGFBR1 LBD Mutants in Cancer / receptor ligand inhibitor activity / positive regulation of chemotaxis / type I transforming growth factor beta receptor binding / response to salt / germ cell migration / positive regulation of mononuclear cell migration / endoderm development / negative regulation of biomineral tissue development / phospholipid homeostasis / negative regulation of cell-cell adhesion mediated by cadherin / negative regulation of myoblast differentiation / positive regulation of vascular permeability / response to cholesterol / oligodendrocyte development / cell-cell junction organization / negative regulation of interleukin-17 production / phosphate-containing compound metabolic process / surfactant homeostasis / transforming growth factor beta binding / deubiquitinase activator activity / sprouting angiogenesis / negative regulation of release of sequestered calcium ion into cytosol / positive regulation of chemokine (C-X-C motif) ligand 2 production / negative regulation of ossification / digestive tract development / aortic valve morphogenesis / RUNX3 regulates CDKN1A transcription / response to vitamin D / face morphogenesis / positive regulation of fibroblast migration / ureteric bud development / neural tube development / positive regulation of regulatory T cell differentiation / positive regulation of peptidyl-tyrosine phosphorylation / Molecules associated with elastic fibres / negative regulation of phagocytosis / positive regulation of epidermal growth factor receptor signaling pathway / negative regulation of neuroblast proliferation / negative regulation of cytokine production / lung alveolus development / Syndecan interactions 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.7 Å | |||||||||
 データ登録者 データ登録者 | Ebenhoch R / Nar H | |||||||||
| 資金援助 | 1件
| |||||||||
 引用 引用 |  ジャーナル: Immunohorizons / 年: 2023 ジャーナル: Immunohorizons / 年: 2023タイトル: Anti-GARP Antibodies Inhibit Release of TGF-β by Regulatory T Cells via Different Modes of Action, but Do Not Influence Their Function In Vitro. 著者: Frederik H Igney / Rebecca Ebenhoch / Felix Schiele / Herbert Nar /  要旨: Regulatory T cells (Treg) play a critical role in controlling immune responses in diseases such as cancer or autoimmunity. Activated Treg express the membrane protein GARP (LRRC32) in complex with ...Regulatory T cells (Treg) play a critical role in controlling immune responses in diseases such as cancer or autoimmunity. Activated Treg express the membrane protein GARP (LRRC32) in complex with the latent form of the immunosuppressive cytokine TGF-β (L-TGF-β). In this study, we confirmed that active TGF-β was generated from its latent form in an integrin-dependent manner and induced TGF-β receptor signaling in activated human Treg. We studied a series of Abs targeting the L-TGF-β/GARP complex with distinct binding modes. We found that TGF-β receptor signaling could be inhibited by anti-TGF-β and by some, but not all, Abs against the L-TGF-β/GARP complex. Cryogenic electron microscopy structures of three L-TGF-β/GARP complex-targeting Abs revealed their distinct epitopes and allowed us to elucidate how they achieve blockade of TGF-β activation. Three different modes of action were identified, including a novel unusual mechanism of a GARP-binding Ab. However, blockade of GARP or TGF-β by Abs did not influence the suppressive activity of human Treg in vitro. We were also not able to confirm a prominent role of GARP in other functions of human Treg, such as FOXP3 induction and Treg stability. These data show that the GARP/TGF-β axis can be targeted pharmacologically in different ways, but further studies are necessary to understand its complexity and to unleash its therapeutic potential. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_16460.map.gz emd_16460.map.gz | 8.3 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-16460-v30.xml emd-16460-v30.xml emd-16460.xml emd-16460.xml | 22.1 KB 22.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_16460.png emd_16460.png | 73 KB | ||
| Filedesc metadata |  emd-16460.cif.gz emd-16460.cif.gz | 7.5 KB | ||
| その他 |  emd_16460_half_map_1.map.gz emd_16460_half_map_1.map.gz emd_16460_half_map_2.map.gz emd_16460_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16460 http://ftp.pdbj.org/pub/emdb/structures/EMD-16460 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16460 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16460 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8c7hMC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_16460.map.gz / 形式: CCP4 / 大きさ: 8.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_16460.map.gz / 形式: CCP4 / 大きさ: 8.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Sharpened map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 これらの図は立方格子座標系で作成されたものです | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: Half map B
| ファイル | emd_16460_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half map B | ||||||||||||
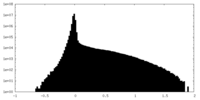
| 投影像・断面図 |
| ||||||||||||
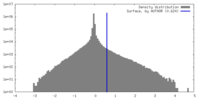
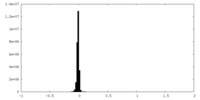
| 密度ヒストグラム |
-ハーフマップ: Half map A
| ファイル | emd_16460_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half map A | ||||||||||||
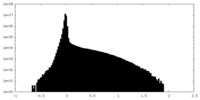
| 投影像・断面図 |
| ||||||||||||
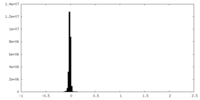
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : latTGF-beta in complex with Fab 28G11
| 全体 | 名称: latTGF-beta in complex with Fab 28G11 |
|---|---|
| 要素 |
|
-超分子 #1: latTGF-beta in complex with Fab 28G11
| 超分子 | 名称: latTGF-beta in complex with Fab 28G11 / タイプ: complex / ID: 1 / 親要素: 0 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Transforming growth factor beta-1
| 分子 | 名称: Transforming growth factor beta-1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 28.531488 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: LSTCKTIDME LVKRKRIEAI RGQILSKLRL ASPPSQGEVP PGPLPEAVLA LYNSTRDRVA GESAEPEPEP EADYYAKEVT RVLMVETHN EIYDKFKQST HSIYMFFNTS ELREAVPEPV LLSRAELRLL RLKLKVEQHV ELYQKYSNNS WRYLSNRLLA P SDSPEWLS ...文字列: LSTCKTIDME LVKRKRIEAI RGQILSKLRL ASPPSQGEVP PGPLPEAVLA LYNSTRDRVA GESAEPEPEP EADYYAKEVT RVLMVETHN EIYDKFKQST HSIYMFFNTS ELREAVPEPV LLSRAELRLL RLKLKVEQHV ELYQKYSNNS WRYLSNRLLA P SDSPEWLS FDVTGVVRQW LSRGGEIEGF RLSAHCSCDS RDNTLQVDIN GFTTGRRGDL ATIHGMNRPF LLLMATPLER AQ HLQSSRH RR UniProtKB: Transforming growth factor beta-1 proprotein |
-分子 #2: Transforming growth factor beta-1
| 分子 | 名称: Transforming growth factor beta-1 / タイプ: protein_or_peptide / ID: 2 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 12.809812 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: ALDTNYCFSS TEKNCCVRQL YIDFRKDLGW KWIHEPKGYH ANFCLGPCPY IWSLDTQYSK VLALYNQHNP GASAAPCCVP QALEPLPIV YYVGRKPKVE QLSNMIVRSC KCS UniProtKB: Transforming growth factor beta-1 proprotein |
-分子 #3: Transforming growth factor beta activator LRRC32
| 分子 | 名称: Transforming growth factor beta activator LRRC32 / タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 65.358488 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: HQDKVPCKMV DKKVSCQVLG LLQVPSVLPP DTETLDLSGN QLRSILASPL GFYTALRHLD LSTNEISFLQ PGAFQALTHL EHLSLAHNR LAMATALSAG GLGPLPRVTS LDLSGNSLYS GLLERLLGEA PSLHTLSLAE NSLTRLTRHT FRDMPALEQL D LHSNVLMD ...文字列: HQDKVPCKMV DKKVSCQVLG LLQVPSVLPP DTETLDLSGN QLRSILASPL GFYTALRHLD LSTNEISFLQ PGAFQALTHL EHLSLAHNR LAMATALSAG GLGPLPRVTS LDLSGNSLYS GLLERLLGEA PSLHTLSLAE NSLTRLTRHT FRDMPALEQL D LHSNVLMD IEDGAFEGLP RLTHLNLSRN SLTCISDFSL QQLRVLDLSC NSIEAFQTAS QPQAEFQLTW LDLRENKLLH FP DLAALPR LIYLNLSNNL IRLPTGPPQD SKGIHAPSEG WSALPLSAPS GNASGRPLSQ LLNLDLSYNE IELIPDSFLE HLT SLCFLN LSRNCLRTFE ARRLGSLPCL MLLDLSHNAL ETLELGARAL GSLRTLLLQG NALRDLPPYT FANLASLQRL NLQG NRVSP CGGPDEPGPS GCVAFSGITS LRSLSLVDNE IELLRAGAFL HTPLTELDLS SNPGLEVATG ALGGLEASLE VLALQ GNGL MVLQVDLPCF ICLKRLNLAE NRLSHLPAWT QAVSLEVLDL RNNSFSLLPG SAMGGLETSL RRLYLQGNPL SCCGNG WLA AQLHQGRVDV DATQDLICRF SSQEEVSLSH VRPEDCEK UniProtKB: Transforming growth factor beta activator LRRC32 |
-分子 #4: 28G11 Fab heavy chain
| 分子 | 名称: 28G11 Fab heavy chain / タイプ: protein_or_peptide / ID: 4 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 24.529211 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: EVQLVQPGAE LRNSGASVKV SCKASGYRFT SYYIDWVRQA PGQGLEWMGR IDPEDGGTKY AQKFQGRVTF TADTSTSTAY VELSSLRSE DTAVYYCARN EWETVVVGDL MYEYEYWGQG TQVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN ...文字列: EVQLVQPGAE LRNSGASVKV SCKASGYRFT SYYIDWVRQA PGQGLEWMGR IDPEDGGTKY AQKFQGRVTF TADTSTSTAY VELSSLRSE DTAVYYCARN EWETVVVGDL MYEYEYWGQG TQVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKRVEPK |
-分子 #5: 28G11 Fab light chain
| 分子 | 名称: 28G11 Fab light chain / タイプ: protein_or_peptide / ID: 5 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 23.091578 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: DIQMTQSPSS LSASLGDRVT ITCQASQSIS SYLAWYQQKP GQAPNILIYG ASRLKTGVPS RFSGSGSGTS FTLTISGLEA EDAGTYYCQ QYASVPVTFG QGTKVELKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...文字列: DIQMTQSPSS LSASLGDRVT ITCQASQSIS SYLAWYQQKP GQAPNILIYG ASRLKTGVPS RFSGSGSGTS FTLTISGLEA EDAGTYYCQ QYASVPVTFG QGTKVELKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 |
|---|---|
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / メッシュ: 300 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 前処理 - タイプ: GLOW DISCHARGE |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 80 % / チャンバー内温度: 4 K / 装置: LEICA EM GP |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 40.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD 最大 デフォーカス(公称値): 2.8000000000000003 µm 最小 デフォーカス(公称値): 0.8 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー






















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)