+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Map of the latTGF-beta 12G2B4 Fab complex | |||||||||
 Map data Map data | Cryo-EM map sharp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fab-complex / GARP / lat-TGF-beta / IMMUNE SYSTEM | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Ebenhoch R / Nar H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Immunohorizons / Year: 2023 Journal: Immunohorizons / Year: 2023Title: Anti-GARP Antibodies Inhibit Release of TGF-β by Regulatory T Cells via Different Modes of Action, but Do Not Influence Their Function In Vitro. Authors: Frederik H Igney / Rebecca Ebenhoch / Felix Schiele / Herbert Nar /  Abstract: Regulatory T cells (Treg) play a critical role in controlling immune responses in diseases such as cancer or autoimmunity. Activated Treg express the membrane protein GARP (LRRC32) in complex with ...Regulatory T cells (Treg) play a critical role in controlling immune responses in diseases such as cancer or autoimmunity. Activated Treg express the membrane protein GARP (LRRC32) in complex with the latent form of the immunosuppressive cytokine TGF-β (L-TGF-β). In this study, we confirmed that active TGF-β was generated from its latent form in an integrin-dependent manner and induced TGF-β receptor signaling in activated human Treg. We studied a series of Abs targeting the L-TGF-β/GARP complex with distinct binding modes. We found that TGF-β receptor signaling could be inhibited by anti-TGF-β and by some, but not all, Abs against the L-TGF-β/GARP complex. Cryogenic electron microscopy structures of three L-TGF-β/GARP complex-targeting Abs revealed their distinct epitopes and allowed us to elucidate how they achieve blockade of TGF-β activation. Three different modes of action were identified, including a novel unusual mechanism of a GARP-binding Ab. However, blockade of GARP or TGF-β by Abs did not influence the suppressive activity of human Treg in vitro. We were also not able to confirm a prominent role of GARP in other functions of human Treg, such as FOXP3 induction and Treg stability. These data show that the GARP/TGF-β axis can be targeted pharmacologically in different ways, but further studies are necessary to understand its complexity and to unleash its therapeutic potential. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16459.map.gz emd_16459.map.gz | 96.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16459-v30.xml emd-16459-v30.xml emd-16459.xml emd-16459.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16459.png emd_16459.png | 68.1 KB | ||
| Filedesc metadata |  emd-16459.cif.gz emd-16459.cif.gz | 3.9 KB | ||
| Others |  emd_16459_half_map_1.map.gz emd_16459_half_map_1.map.gz emd_16459_half_map_2.map.gz emd_16459_half_map_2.map.gz | 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16459 http://ftp.pdbj.org/pub/emdb/structures/EMD-16459 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16459 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16459 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16459.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16459.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map sharp | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
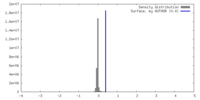
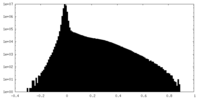
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM half map B
| File | emd_16459_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
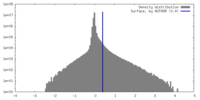
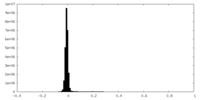
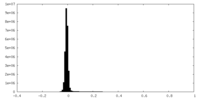
| Density Histograms |
-Half map: Cryo-EM half map A
| File | emd_16459_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
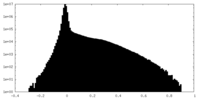
| Density Histograms |
- Sample components
Sample components
-Entire : latTGF-beta in complex with Fab 28G11
| Entire | Name: latTGF-beta in complex with Fab 28G11 |
|---|---|
| Components |
|
-Supramolecule #1: latTGF-beta in complex with Fab 28G11
| Supramolecule | Name: latTGF-beta in complex with Fab 28G11 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 4 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 613675 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)