[English] 日本語
 Yorodumi
Yorodumi- EMDB-1639: Proteome organization in a genome-reduced bacterium -RNA-polymera... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1639 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Proteome organization in a genome-reduced bacterium -RNA-polymeraseof Mycoplasma pneumoniae - | |||||||||
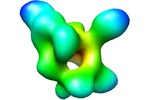
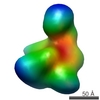
 Map data Map data | RNA-polymerase from Mycoplasma pneumoniae | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA-polymerase / Mycoplasma pneumoniae / single particle | |||||||||
| Biological species |  Mycoplasma pneumoniae (Filterable agent of primary atypical pneumonia) Mycoplasma pneumoniae (Filterable agent of primary atypical pneumonia) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Kuhner S / vanNoort V / Betts MJ / Leo-Macias A / Batisse C / Rode M / Yamada T / Maier T / Bader S / Beltran-Alvarez P ...Kuhner S / vanNoort V / Betts MJ / Leo-Macias A / Batisse C / Rode M / Yamada T / Maier T / Bader S / Beltran-Alvarez P / Castano-Diez D / Chen W-H / Devos D / Guell Cargol M / Norambuena T / Racke I / Rybin V / Schmidt A / Yus E / Aebersold R / Herrmann R / Bottcher B / Frangakis AS / Russell RB / Serrano L / Bork P / Gavin A-C | |||||||||
 Citation Citation |  Journal: Science / Year: 2009 Journal: Science / Year: 2009Title: Proteome organization in a genome-reduced bacterium. Authors: Sebastian Kühner / Vera van Noort / Matthew J Betts / Alejandra Leo-Macias / Claire Batisse / Michaela Rode / Takuji Yamada / Tobias Maier / Samuel Bader / Pedro Beltran-Alvarez / Daniel ...Authors: Sebastian Kühner / Vera van Noort / Matthew J Betts / Alejandra Leo-Macias / Claire Batisse / Michaela Rode / Takuji Yamada / Tobias Maier / Samuel Bader / Pedro Beltran-Alvarez / Daniel Castaño-Diez / Wei-Hua Chen / Damien Devos / Marc Güell / Tomas Norambuena / Ines Racke / Vladimir Rybin / Alexander Schmidt / Eva Yus / Ruedi Aebersold / Richard Herrmann / Bettina Böttcher / Achilleas S Frangakis / Robert B Russell / Luis Serrano / Peer Bork / Anne-Claude Gavin /  Abstract: The genome of Mycoplasma pneumoniae is among the smallest found in self-replicating organisms. To study the basic principles of bacterial proteome organization, we used tandem affinity purification- ...The genome of Mycoplasma pneumoniae is among the smallest found in self-replicating organisms. To study the basic principles of bacterial proteome organization, we used tandem affinity purification-mass spectrometry (TAP-MS) in a proteome-wide screen. The analysis revealed 62 homomultimeric and 116 heteromultimeric soluble protein complexes, of which the majority are novel. About a third of the heteromultimeric complexes show higher levels of proteome organization, including assembly into larger, multiprotein complex entities, suggesting sequential steps in biological processes, and extensive sharing of components, implying protein multifunctionality. Incorporation of structural models for 484 proteins, single-particle electron microscopy, and cellular electron tomograms provided supporting structural details for this proteome organization. The data set provides a blueprint of the minimal cellular machinery required for life. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1639.map.gz emd_1639.map.gz | 22.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1639-v30.xml emd-1639-v30.xml emd-1639.xml emd-1639.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1639.png 1639.png | 285.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1639 http://ftp.pdbj.org/pub/emdb/structures/EMD-1639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1639 | HTTPS FTP |
-Validation report
| Summary document |  emd_1639_validation.pdf.gz emd_1639_validation.pdf.gz | 183.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1639_full_validation.pdf.gz emd_1639_full_validation.pdf.gz | 182.8 KB | Display | |
| Data in XML |  emd_1639_validation.xml.gz emd_1639_validation.xml.gz | 4.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1639 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1639 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1639 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1639 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1639.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1639.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RNA-polymerase from Mycoplasma pneumoniae | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RNA-polymerase of Mycoplasma pneumoniae
| Entire | Name: RNA-polymerase of Mycoplasma pneumoniae |
|---|---|
| Components |
|
-Supramolecule #1000: RNA-polymerase of Mycoplasma pneumoniae
| Supramolecule | Name: RNA-polymerase of Mycoplasma pneumoniae / type: sample / ID: 1000 / Details: Sample was fixed following the GRAFIX protocol / Number unique components: 1 |
|---|
-Macromolecule #1: RNA-polymerase
| Macromolecule | Name: RNA-polymerase / type: protein_or_peptide / ID: 1 / Name.synonym: Polymerase / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Mycoplasma pneumoniae (Filterable agent of primary atypical pneumonia) Mycoplasma pneumoniae (Filterable agent of primary atypical pneumonia) |
| Recombinant expression | Organism:  Mycoplasma pneumoniae (Filterable agent of primary atypical pneumonia) Mycoplasma pneumoniae (Filterable agent of primary atypical pneumonia) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 50mM Hepes, 20% glycerol, 0.075% glutaraldehyde, 100mM NaCl, 1.5 mM MgCl2 |
|---|---|
| Staining | Type: NEGATIVE Details: Grids were prepared by sandwich negative stain the sample was adsorbed to carbon on mica and floated on 1 % uranyl acetate the carbon was picked up with an uncoated grid then a second piece ...Details: Grids were prepared by sandwich negative stain the sample was adsorbed to carbon on mica and floated on 1 % uranyl acetate the carbon was picked up with an uncoated grid then a second piece of carbon, which was also floated on 1% uranyl acetate, was picked up with the same grid sandwiching the sample between the two layers of carbon |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Corrected at 200000 times magnification on graininess of carbon |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Digitization - Sampling interval: 14.22 µm / Number real images: 96 Details: Images were recorded on CCD, no scanning sampling step size was adjusted to calibrated image size Bits/pixel: 12 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal magnification: 27500 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC, SPIDER, EMAN Details: Spider option BP 32F Back Projection - 3D, Sampled, Interpolated in Fourier space Number images used: 5324 |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















