[English] 日本語
 Yorodumi
Yorodumi- EMDB-16281: Cryo-EM structure of the RAF activating complex KSR-MEK-CNK-HYP -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the RAF activating complex KSR-MEK-CNK-HYP | ||||||||||||
 Map data Map data | Final Map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Kinase suppressor of Ras (KSR) / Dual specificity mitogen-activated protein kinase kinase (MEK) / Connector enhancer of KSR (CNK) / Protein Aveugle (AVE) / Hyphen protein (HYP) / protein complex / SIGNALING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationhemocyte differentiation / MAPK3 (ERK1) activation / MAPK1 (ERK2) activation / Frs2-mediated activation / Signal transduction by L1 / Negative feedback regulation of MAPK pathway / terminal region determination / RAF activation / Phosphorylation of CI / compound eye cone cell differentiation ...hemocyte differentiation / MAPK3 (ERK1) activation / MAPK1 (ERK2) activation / Frs2-mediated activation / Signal transduction by L1 / Negative feedback regulation of MAPK pathway / terminal region determination / RAF activation / Phosphorylation of CI / compound eye cone cell differentiation / Phosphorylation of SMO / terminal branching, open tracheal system / torso signaling pathway / photoreceptor cell development / tracheal outgrowth, open tracheal system / Phosphorylation of PER and TIM / compound eye photoreceptor cell differentiation / MAP2K and MAPK activation / eye photoreceptor cell differentiation / epithelial cell migration, open tracheal system / R7 cell fate commitment / sevenless signaling pathway / imaginal disc-derived wing vein specification / border follicle cell migration / imaginal disc-derived wing morphogenesis / anterior/posterior axis specification, embryo / mitogen-activated protein kinase kinase / cellular response to X-ray / MAP-kinase scaffold activity / mitogen-activated protein kinase kinase kinase binding / mitotic DNA replication checkpoint signaling / dorsal/ventral pattern formation / positive regulation of Ras protein signal transduction / mitotic G2 DNA damage checkpoint signaling / MAP kinase kinase activity / fibroblast growth factor receptor signaling pathway / vascular endothelial growth factor receptor signaling pathway / condensed chromosome / enzyme regulator activity / visual perception / ERK1 and ERK2 cascade / cell surface receptor protein tyrosine kinase signaling pathway / determination of adult lifespan / receptor tyrosine kinase binding / cytoplasmic side of plasma membrane / kinase binding / epidermal growth factor receptor signaling pathway / apical part of cell / MAPK cascade / insulin receptor signaling pathway / cell cortex / protein tyrosine kinase activity / scaffold protein binding / defense response to virus / Ras protein signal transduction / positive regulation of ERK1 and ERK2 cascade / protein kinase activity / protein serine kinase activity / protein serine/threonine kinase activity / ATP binding / metal ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | ||||||||||||
 Authors Authors | Maisonneuve P / Fronzes R / Sicheri F | ||||||||||||
| Funding support |  Canada, Canada,  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: The CNK-HYP scaffolding complex promotes RAF activation by enhancing KSR-MEK interaction. Authors: Pierre Maisonneuve / Malha Sahmi / Fanny Bergeron-Labrecque / Xianjie Iris Ma / Juliette Queguiner / Geneviève Arseneault / Martin Lefrançois / Igor Kurinov / Rémi Fronzes / Frank Sicheri / Marc Therrien /    Abstract: The RAS-MAPK pathway regulates cell proliferation, differentiation and survival, and its dysregulation is associated with cancer development. The pathway minimally comprises the small GTPase RAS and ...The RAS-MAPK pathway regulates cell proliferation, differentiation and survival, and its dysregulation is associated with cancer development. The pathway minimally comprises the small GTPase RAS and the kinases RAF, MEK and ERK. Activation of RAF by RAS is notoriously intricate and remains only partially understood. There are three RAF isoforms in mammals (ARAF, BRAF and CRAF) and two related pseudokinases (KSR1 and KSR2). RAS-mediated activation of RAF depends on an allosteric mechanism driven by the dimerization of its kinase domain. Recent work on human RAFs showed that MEK binding to KSR1 promotes KSR1-BRAF heterodimerization, which leads to the phosphorylation of free MEK molecules by BRAF. Similar findings were made with the single Drosophila RAF homolog. Here we show that the fly scaffold proteins CNK and HYP stabilize the KSR-MEK interaction, which in turn enhances RAF-KSR heterodimerization and RAF activation. The cryogenic electron microscopy structure of the minimal KSR-MEK-CNK-HYP complex reveals a ring-like arrangement of the CNK-HYP complex allowing CNK to simultaneously engage KSR and MEK, thus stabilizing the binary interaction. Together, these results illuminate how CNK contributes to RAF activation by stimulating the allosteric function of KSR and highlight the diversity of mechanisms impacting RAF dimerization as well as the regulatory potential of the KSR-MEK interaction. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16281.map.gz emd_16281.map.gz | 167.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16281-v30.xml emd-16281-v30.xml emd-16281.xml emd-16281.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16281.png emd_16281.png | 51.3 KB | ||
| Masks |  emd_16281_msk_1.map emd_16281_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16281.cif.gz emd-16281.cif.gz | 7.3 KB | ||
| Others |  emd_16281_half_map_1.map.gz emd_16281_half_map_1.map.gz emd_16281_half_map_2.map.gz emd_16281_half_map_2.map.gz | 165.2 MB 165.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16281 http://ftp.pdbj.org/pub/emdb/structures/EMD-16281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16281 | HTTPS FTP |
-Related structure data
| Related structure data |  8bw9MC  8bw8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16281.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16281.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
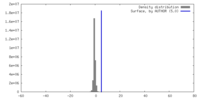
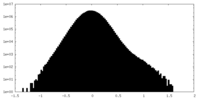
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16281_msk_1.map emd_16281_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
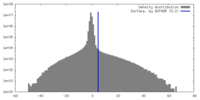
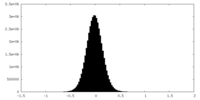
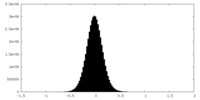
| Density Histograms |
-Half map: Half-map B
| File | emd_16281_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map A
| File | emd_16281_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Quaternary complex of the kinase domains of KSR and MEK bound to ...
| Entire | Name: Quaternary complex of the kinase domains of KSR and MEK bound to the scaffolding complex CNK-HYP |
|---|---|
| Components |
|
-Supramolecule #1: Quaternary complex of the kinase domains of KSR and MEK bound to ...
| Supramolecule | Name: Quaternary complex of the kinase domains of KSR and MEK bound to the scaffolding complex CNK-HYP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: Mg2+/ANP and Trametinib ligands bound to MEK |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 132 KDa |
-Macromolecule #1: Protein aveugle
| Macromolecule | Name: Protein aveugle / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.143911 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMDPGEETI NSTQNKTRTK TTRPKAVYLW TVSDVLKWYR RHCGEYTQYE QLFAQHDITG RALLRITDSS LQRMGVTDNR DREAIWREI VKQRLKTDIM EIRDMERLNI Y UniProtKB: Protein aveugle |
-Macromolecule #2: Connector enhancer of KSR protein CNK
| Macromolecule | Name: Connector enhancer of KSR protein CNK / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.534203 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMDPMAYIN IAEWTPDQVT DWIKGLDESM KGYLYEFSKQ EIGGRALLNI RPYELENLGM LRIGHQEIVL EAVENLRNFH YHLKNDNLQ FMALHVATAA KNLHRELARN HAESTKIDTR ILHDITRTIA TLKPLVGSLE RTPFRKQEMY REYCGNVLKC G LELATIAH ...String: GAMDPMAYIN IAEWTPDQVT DWIKGLDESM KGYLYEFSKQ EIGGRALLNI RPYELENLGM LRIGHQEIVL EAVENLRNFH YHLKNDNLQ FMALHVATAA KNLHRELARN HAESTKIDTR ILHDITRTIA TLKPLVGSLE RTPFRKQEMY REYCGNVLKC G LELATIAH RDRFALQPVP AIRQSAERLE NLANFVIQDI SDPMVLQPAS LNLVTLKKRE SELGFNIESS YNGIHRVTDI KY NSPAHNS GKIEDGDEIV QINYQTVVGW QHRTVLEHLR EALPDVVLTV KKRPKHTKMF GQIYMQPYRL PSKKRNMAAR WAA QMPSPR AAFLTLDTE UniProtKB: Connector enhancer of KSR protein CNK |
-Macromolecule #3: Dual specificity mitogen-activated protein kinase kinase dSOR1
| Macromolecule | Name: Dual specificity mitogen-activated protein kinase kinase dSOR1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: mitogen-activated protein kinase kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.926496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKNKLNLVL PPVNTEATVA AATVAPTPPF KTPSGTDTHS LLGKPKTSID ALTETLEGLD MGDTERKRIK MFLSQKEKIG ELSDEDLEK LGELGSGNGG VVMKVRHTHT HLIMARKLIH LEVKPAIKKQ ILRELKVLHE CNFPHIVGFY GAFYSDGEIS I CMEYMDGG ...String: MSKNKLNLVL PPVNTEATVA AATVAPTPPF KTPSGTDTHS LLGKPKTSID ALTETLEGLD MGDTERKRIK MFLSQKEKIG ELSDEDLEK LGELGSGNGG VVMKVRHTHT HLIMARKLIH LEVKPAIKKQ ILRELKVLHE CNFPHIVGFY GAFYSDGEIS I CMEYMDGG SLDLILKRAG RIPESILGRI TLAVLKGLSY LRDNHAIIHR DVKPSNILVN SSGEIKICDF GVSGQLIDSM AN SFVGTRS YMSPERLQGT HYSVQSDIWS LGLSLVEMAI GMYPIPPPNT ATLESIFADN AEESGQPTDE PRAMAIFELL DYI VNEPPP KLEHKIFSTE FKDFVDICLK KQPDERADLK TLLSHPWIRK AELEEVDISG WVCKTMDLPP STPKRNTSPN UniProtKB: Dual specificity mitogen-activated protein kinase kinase dSOR1 |
-Macromolecule #4: KSR
| Macromolecule | Name: KSR / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.474898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGRTEDGDSG QWRQNSISLK EWDIPYGDLL LLERIGQGRF GTVHRALWHG DVAVKLLNED YLQDEHMLET FRSEVANFKN TRHENLVLF MGACMNPPYL AIVTSLCKGN TLYTYIHQRR EKFAMNRTLL IAQQIAQGMG YLHAREIIHK DLRTKNIFIE N GKVIITDF ...String: GGRTEDGDSG QWRQNSISLK EWDIPYGDLL LLERIGQGRF GTVHRALWHG DVAVKLLNED YLQDEHMLET FRSEVANFKN TRHENLVLF MGACMNPPYL AIVTSLCKGN TLYTYIHQRR EKFAMNRTLL IAQQIAQGMG YLHAREIIHK DLRTKNIFIE N GKVIITDF GLFSSTKLLY CDMGLGVPHN WLCYLAPELI RALQPEKPRG ECLEFTPYSD VYSFGTVWYE LICGEFTFKD QP AESIIWQ VGRGMKQSLA NLQSGRDVKD LLMLCWTYEK EHRPQFARLL SLLEHLPKKR LARSPSHPVN LSRSAESVF UniProtKB: KSR |
-Macromolecule #5: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #6: Trametinib
| Macromolecule | Name: Trametinib / type: ligand / ID: 6 / Number of copies: 1 / Formula: QOM |
|---|---|
| Molecular weight | Theoretical: 615.395 Da |
| Chemical component information |  ChemComp-QOM: |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.79 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: AMPPNP 25mM Trametinib 0.05mM | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.00026000000000000003 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: 3.5 sec blot time 1 sec drain time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 44 / Average electron dose: 1.15 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 26.398 µm / Nominal defocus min: 5.212 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.32 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 141531 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)