[English] 日本語
 Yorodumi
Yorodumi- EMDB-1614: Globular tetramers of beta-2-microglobulin assemble into elaborat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1614 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Globular tetramers of beta-2-microglobulin assemble into elaborate amyloid fibrils | |||||||||
 Map data Map data | Surface view of an B-type map from beta-2-microglobulin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | protein misfolding / amyloid / cryo-EM / 3D reconstruction / STEM | |||||||||
| Function / homology | Beta-2-Microglobulin / MHC class I protein complex Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 25.0 Å | |||||||||
 Authors Authors | White HE / Hodgkinson JL / Jahn TR / Cohen-Krausz S / Gosal WS / Muller S / Orlova EV / Radford SE / Saibil HR | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2009 Journal: J Mol Biol / Year: 2009Title: Globular tetramers of beta(2)-microglobulin assemble into elaborate amyloid fibrils. Authors: Helen E White / Julie L Hodgkinson / Thomas R Jahn / Sara Cohen-Krausz / Walraj S Gosal / Shirley Müller / Elena V Orlova / Sheena E Radford / Helen R Saibil /  Abstract: Amyloid fibrils are ordered polymers in which constituent polypeptides adopt a non-native fold. Despite their importance in degenerative human diseases, the overall structure of amyloid fibrils ...Amyloid fibrils are ordered polymers in which constituent polypeptides adopt a non-native fold. Despite their importance in degenerative human diseases, the overall structure of amyloid fibrils remains unknown. High-resolution studies of model peptide assemblies have identified residues forming cross-beta-strands and have revealed some details of local beta-strand packing. However, little is known about the assembly contacts that define the fibril architecture. Here we present a set of three-dimensional structures of amyloid fibrils formed from full-length beta(2)-microglobulin, a 99-residue protein involved in clinical amyloidosis. Our cryo-electron microscopy maps reveal a hierarchical fibril structure built from tetrameric units of globular density, with at least three different subunit interfaces in this homopolymeric assembly. These findings suggest a more complex superstructure for amyloid than hitherto suspected and prompt a re-evaluation of the defining features of the amyloid fold. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1614.map.gz emd_1614.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1614-v30.xml emd-1614-v30.xml emd-1614.xml emd-1614.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  1614.tif 1614.tif | 737.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1614 http://ftp.pdbj.org/pub/emdb/structures/EMD-1614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1614 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1614.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1614.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Surface view of an B-type map from beta-2-microglobulin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
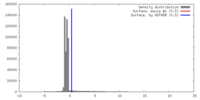
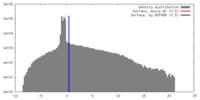
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : beta-2-microglobulin
| Entire | Name: beta-2-microglobulin |
|---|---|
| Components |
|
-Supramolecule #1000: beta-2-microglobulin
| Supramolecule | Name: beta-2-microglobulin / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 10 KDa |
-Macromolecule #1: beta-2-microglobulin
| Macromolecule | Name: beta-2-microglobulin / type: protein_or_peptide / ID: 1 / Name.synonym: beta-2-microglobulin / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: plasma Homo sapiens (human) / synonym: Human / Location in cell: plasma |
| Molecular weight | Experimental: 10 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: MHC class I protein complex / InterPro: Beta-2-Microglobulin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 2.5 / Details: 25 mM sodium phosphate, 25 mM sodium acetate |
| Grid | Details: holey carbon grids |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: manual plunger |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 100 K |
| Details | Low dose |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 85 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder: side entry / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 52.5 Å Applied symmetry - Helical parameters - Δ&Phi: 7.01 ° Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER Details: Maps were calculated for each class as each class had a different pitch. |
|---|---|
| CTF correction | Details: Phase flipping |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)