[English] 日本語
 Yorodumi
Yorodumi- EMDB-1603: 12 Angstrom resolution cryo-electron microscopy reconstruction of... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 12 Angstrom resolution cryo-electron microscopy reconstruction of a recombinant active ribonucleoprotein particle of influenza virus (9-fold symmetrized). | |||||||||
 Map data Map data | Density map of a influenza virus recombinant active RNP refined with 9-fold symmetry. Density corresponding to the polymerase has been smeared out due to the symmetrization. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza / RNA viruses / nucleoprotein / RNA | |||||||||
| Function / homology |  Function and homology information Function and homology informationcRNA Synthesis / Assembly of Viral Components at the Budding Site / Influenza Infection / Fusion of the Influenza Virion to the Host Cell Endosome / Release / Budding / Packaging of Eight RNA Segments / Uncoating of the Influenza Virion / Entry of Influenza Virion into Host Cell via Endocytosis / Viral RNP Complexes in the Host Cell Nucleus ...cRNA Synthesis / Assembly of Viral Components at the Budding Site / Influenza Infection / Fusion of the Influenza Virion to the Host Cell Endosome / Release / Budding / Packaging of Eight RNA Segments / Uncoating of the Influenza Virion / Entry of Influenza Virion into Host Cell via Endocytosis / Viral RNP Complexes in the Host Cell Nucleus / vRNA Synthesis / Transport of Ribonucleoproteins into the Host Nucleus / NEP/NS2 Interacts with the Cellular Export Machinery / Viral Messenger RNA Synthesis / vRNP Assembly / helical viral capsid / Viral mRNA Translation / viral penetration into host nucleus / host cell / viral nucleocapsid / ribonucleoprotein complex / symbiont entry into host cell / host cell nucleus / structural molecule activity / RNA binding / extracellular region / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Influenza A virus (A/Victoria/3/1975(H3N2)) Influenza A virus (A/Victoria/3/1975(H3N2)) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 12.0 Å | |||||||||
 Authors Authors | Coloma R / Valpuesta JM / Arranz R / Carrascosa JL / Ortin J / Martin-Benito J | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2009 Journal: PLoS Pathog / Year: 2009Title: The structure of a biologically active influenza virus ribonucleoprotein complex. Authors: Rocío Coloma / José M Valpuesta / Rocío Arranz / José L Carrascosa / Juan Ortín / Jaime Martín-Benito /  Abstract: The influenza viruses contain a segmented, single-stranded RNA genome of negative polarity. Each RNA segment is encapsidated by the nucleoprotein and the polymerase complex into ribonucleoprotein ...The influenza viruses contain a segmented, single-stranded RNA genome of negative polarity. Each RNA segment is encapsidated by the nucleoprotein and the polymerase complex into ribonucleoprotein particles (RNPs), which are responsible for virus transcription and replication. Despite their importance, information about the structure of these RNPs is scarce. We have determined the three-dimensional structure of a biologically active recombinant RNP by cryo-electron microscopy. The structure shows a nonameric nucleoprotein ring (at 12 Angstrom resolution) with two monomers connected to the polymerase complex (at 18 Angstrom resolution). Docking the atomic structures of the nucleoprotein and polymerase domains, as well as mutational analyses, has allowed us to define the interactions between the functional elements of the RNP and to propose the location of the viral RNA. Our results provide the first model for a functional negative-stranded RNA virus ribonucleoprotein complex. The structure reported here will serve as a framework to generate a quasi-atomic model of the molecular machine responsible for viral RNA synthesis and to test new models for virus RNA replication and transcription. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1603.map.gz emd_1603.map.gz | 14.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1603-v30.xml emd-1603-v30.xml emd-1603.xml emd-1603.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  1603.png 1603.png | 214.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1603 http://ftp.pdbj.org/pub/emdb/structures/EMD-1603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1603 | HTTPS FTP |
-Related structure data
| Related structure data |  2wfsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1603.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1603.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density map of a influenza virus recombinant active RNP refined with 9-fold symmetry. Density corresponding to the polymerase has been smeared out due to the symmetrization. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||
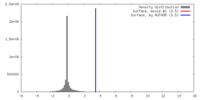
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : influenza virus ribonucleoprotein particle from INFLUENZA A VIRUS...
| Entire | Name: influenza virus ribonucleoprotein particle from INFLUENZA A VIRUS STRAIN A/VICTORIA/3/75 (H3N1 AFRICAN GREEN MONKEY |
|---|---|
| Components |
|
-Supramolecule #1000: influenza virus ribonucleoprotein particle from INFLUENZA A VIRUS...
| Supramolecule | Name: influenza virus ribonucleoprotein particle from INFLUENZA A VIRUS STRAIN A/VICTORIA/3/75 (H3N1 AFRICAN GREEN MONKEY type: sample / ID: 1000 Oligomeric state: Nine monomers of nucleoprotein bound to a single stranded RNA Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: nucleoprotein
| Macromolecule | Name: nucleoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: nucleoprotein / Number of copies: 9 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Victoria/3/1975(H3N2)) / Location in cell: nucleus Influenza A virus (A/Victoria/3/1975(H3N2)) / Location in cell: nucleus |
| Molecular weight | Theoretical: 56 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
-Macromolecule #2: RNA
| Macromolecule | Name: RNA / type: rna / ID: 2 / Name.synonym: RNA / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Victoria/3/1975(H3N2)) Influenza A virus (A/Victoria/3/1975(H3N2)) |
| Molecular weight | Theoretical: 82 KDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: 50mM Tris-HCl,100mM KCl,5mM MgCl2,0.5% Igepal,150mM imidazole |
|---|---|
| Staining | Type: NEGATIVE Details: Samples were applied to one side of a holey carbon grid, blotted and plunged into liquid ethane |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA PLUNGER / Details: Vitrification instrument: Leica plunger |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 159 / Details: downsampling factor 2. / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each plate |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C9 (9 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 12.0 Å / Resolution method: FSC 0.33 CUT-OFF / Software - Name: spider / Number images used: 9571 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: SITUS |
| Details | PDBEntryID_givenInChain. Protocol: Rigid Body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Volumetric |
| Output model |  PDB-2wfs: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)