+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T3 SAM lyase in complex with S-adenosylmethionine synthase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SAM lyase / complex / LYASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationS-adenosyl-L-methionine lyase / S-adenosyl-L-methionine lyase activity / methionine adenosyltransferase / methionine adenosyltransferase activity / S-adenosylmethionine biosynthetic process / S-adenosylmethionine cycle / potassium ion binding / one-carbon metabolic process / transferase activity / magnesium ion binding ...S-adenosyl-L-methionine lyase / S-adenosyl-L-methionine lyase activity / methionine adenosyltransferase / methionine adenosyltransferase activity / S-adenosylmethionine biosynthetic process / S-adenosylmethionine cycle / potassium ion binding / one-carbon metabolic process / transferase activity / magnesium ion binding / ATP binding / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Enterobacteria phage T3 (virus) / Enterobacteria phage T3 (virus) /  | |||||||||
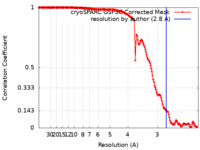
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Triguis S / Selmer M | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase. Authors: Aleksandr Andriianov / Silvia Trigüis / Alena Drobiazko / Nicolas Sierro / Nikolai V Ivanov / Maria Selmer / Konstantin Severinov / Artem Isaev /     Abstract: Bacteriophage T3 encodes a SAMase that, through cleavage of S-adenosyl methionine (SAM), circumvents the SAM-dependent type I restriction-modification (R-M) defense. We show that SAMase also allows ...Bacteriophage T3 encodes a SAMase that, through cleavage of S-adenosyl methionine (SAM), circumvents the SAM-dependent type I restriction-modification (R-M) defense. We show that SAMase also allows T3 to evade the BREX defense. Although SAM depletion weakly affects BREX methylation, it completely inhibits the defensive function of BREX, suggesting that SAM could be a co-factor for BREX-mediated exclusion of phage DNA, similar to its anti-defense role in type I R-M. The anti-BREX activity of T3 SAMase is mediated not just by enzymatic degradation of SAM but also by direct inhibition of MetK, the host SAM synthase. We present a 2.8 Å cryoelectron microscopy (cryo-EM) structure of the eight-subunit T3 SAMase-MetK complex. Structure-guided mutagenesis reveals that this interaction stabilizes T3 SAMase in vivo, further stimulating its anti-BREX activity. This work provides insights in the versatility of bacteriophage counterdefense mechanisms and highlights the role of SAM as a co-factor of diverse bacterial immunity systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15953.map.gz emd_15953.map.gz | 212.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15953-v30.xml emd-15953-v30.xml emd-15953.xml emd-15953.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15953_fsc.xml emd_15953_fsc.xml | 17.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15953.png emd_15953.png | 81 KB | ||
| Masks |  emd_15953_msk_1.map emd_15953_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Others |  emd_15953_half_map_1.map.gz emd_15953_half_map_1.map.gz emd_15953_half_map_2.map.gz emd_15953_half_map_2.map.gz | 390.7 MB 390.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15953 http://ftp.pdbj.org/pub/emdb/structures/EMD-15953 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15953 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15953 | HTTPS FTP |
-Validation report
| Summary document |  emd_15953_validation.pdf.gz emd_15953_validation.pdf.gz | 734.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15953_full_validation.pdf.gz emd_15953_full_validation.pdf.gz | 734.2 KB | Display | |
| Data in XML |  emd_15953_validation.xml.gz emd_15953_validation.xml.gz | 24.8 KB | Display | |
| Data in CIF |  emd_15953_validation.cif.gz emd_15953_validation.cif.gz | 32.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15953 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15953 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15953 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15953 | HTTPS FTP |
-Related structure data
| Related structure data |  8bb1MC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15953.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15953.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1125 Å | ||||||||||||||||||||||||||||||||||||
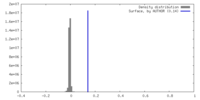
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15953_msk_1.map emd_15953_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
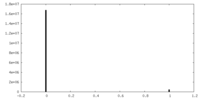
| Density Histograms |
-Half map: #1
| File | emd_15953_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
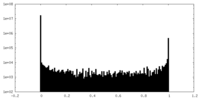
| Density Histograms |
-Half map: #2
| File | emd_15953_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
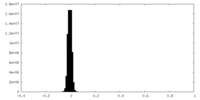
| Density Histograms |
- Sample components
Sample components
-Entire : T3 SAM lyase in complex with E. coli S-adenosylmethionine synthase.
| Entire | Name: T3 SAM lyase in complex with E. coli S-adenosylmethionine synthase. |
|---|---|
| Components |
|
-Supramolecule #1: T3 SAM lyase in complex with E. coli S-adenosylmethionine synthase.
| Supramolecule | Name: T3 SAM lyase in complex with E. coli S-adenosylmethionine synthase. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: T3 SAM lyase was recombinantly expressed in E. coli Top10 and co-purified in complex with S-adenosylmethionine synthase from the host. |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T3 (virus) Enterobacteria phage T3 (virus) |
| Molecular weight | Theoretical: 239 KDa |
-Supramolecule #2: S-adenosylmethionine synthase in complex with T3 SAM lyase.
| Supramolecule | Name: S-adenosylmethionine synthase in complex with T3 SAM lyase. type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 Details: T3 SAM lyase was recombinantly expressed in E. coli Top10 and co-purified in complex with S-adenosylmethionine synthase from the host. |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: T3 SAM lyase in complex with S-adenosylmethionine synthase.
| Supramolecule | Name: T3 SAM lyase in complex with S-adenosylmethionine synthase. type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#2 Details: T3 SAM lyase was recombinantly expressed in E. coli Top10 and co-purified in complex with S-adenosylmethionine synthase from the host. |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T3 (virus) Enterobacteria phage T3 (virus) |
-Macromolecule #1: S-adenosylmethionine synthase
| Macromolecule | Name: S-adenosylmethionine synthase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: methionine adenosyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.998508 KDa |
| Sequence | String: MAKHLFTSES VSEGHPDKIA DQISDAVLDA ILEQDPKARV ACETYVKTGM VLVGGEITTS AWVDIEEITR NTVREIGYVH SDMGFDANS CAVLSAIGKQ SPDINQGVDR ADPLEQGAGD QGLMFGYATN ETDVLMPAPI TYAHRLVQRQ AEVRKNGTLP W LRPDAKSQ ...String: MAKHLFTSES VSEGHPDKIA DQISDAVLDA ILEQDPKARV ACETYVKTGM VLVGGEITTS AWVDIEEITR NTVREIGYVH SDMGFDANS CAVLSAIGKQ SPDINQGVDR ADPLEQGAGD QGLMFGYATN ETDVLMPAPI TYAHRLVQRQ AEVRKNGTLP W LRPDAKSQ VTFQYDDGKI VGIDAVVLST QHSEEIDQKS LQEAVMEEII KPILPAEWLT SATKFFINPT GRFVIGGPMG DC GLTGRKI IVDTYGGMAR HGGGAFSGKD PSKVDRSAAY AARYVAKNIV AAGLADRCEI QVSYAIGVAE PTSIMVETFG TEK VPSEQL TLLVREFFDL RPYGLIQMLD LLHPIYKETA AYGHFGREHF PWEKTDKAQL LRDAAGLK UniProtKB: S-adenosylmethionine synthase |
-Macromolecule #2: S-Adenosylmethionine lyase
| Macromolecule | Name: S-Adenosylmethionine lyase / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: ec: 2.5.1.4 |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T3 (virus) Enterobacteria phage T3 (virus) |
| Molecular weight | Theoretical: 17.863264 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIFTKEPANV FYVLVSAFRS NLCDEVNMSR HRHMVSTLRA APGLYGSVES TDLTGCYREA ISSAPTEEKT VRVRCKDKAQ ALNVARLAC NEWEQDCVLV YKSQTHTAGL VYAKGIDGYK AERLPGSFQE VPKGAPLQGC FTIDEFGRRW QVQHHHHHH UniProtKB: S-Adenosylmethionine lyase |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 24 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.125 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting for 3 seconds before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 5447 / Average exposure time: 3.0 sec. / Average electron dose: 47.75 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.1 µm / Calibrated defocus min: 0.35000000000000003 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)