+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Drosophila melanogaster complex I in the Cracked state (Dm3) | |||||||||
 Map data Map data | Globally sharpened consensus map generated using RELION Postprocess | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
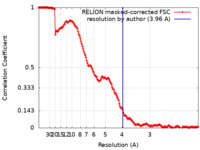
| Method | single particle reconstruction / cryo EM / Resolution: 3.96 Å | |||||||||
 Authors Authors | Agip AA / Chung I / Sanchez-Martinez A / Whitworth AJ / Hirst J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Cryo-EM structures of mitochondrial respiratory complex I from . Authors: Ahmed-Noor A Agip / Injae Chung / Alvaro Sanchez-Martinez / Alexander J Whitworth / Judy Hirst /  Abstract: Respiratory complex I powers ATP synthesis by oxidative phosphorylation, exploiting the energy from NADH oxidation by ubiquinone to drive protons across an energy-transducing membrane. is a ...Respiratory complex I powers ATP synthesis by oxidative phosphorylation, exploiting the energy from NADH oxidation by ubiquinone to drive protons across an energy-transducing membrane. is a candidate model organism for complex I due to its high evolutionary conservation with the mammalian enzyme, well-developed genetic toolkit, and complex physiology for studies in specific cell types and tissues. Here, we isolate complex I from and determine its structure, revealing a 43-subunit assembly with high structural homology to its 45-subunit mammalian counterpart, including a hitherto unknown homologue to subunit NDUFA3. The major conformational state of the enzyme is the mammalian-type 'ready-to-go' active resting state, with a fully ordered and enclosed ubiquinone-binding site, but a subtly altered global conformation related to changes in subunit ND6. The mammalian-type 'deactive' pronounced resting state is not observed: in two minor states, the ubiquinone-binding site is unchanged, but a deactive-type π-bulge is present in ND6-TMH3. Our detailed structural knowledge of complex I provides a foundation for new approaches to disentangle mechanisms of complex I catalysis and regulation in bioenergetics and physiology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15938.map.gz emd_15938.map.gz | 317.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15938-v30.xml emd-15938-v30.xml emd-15938.xml emd-15938.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15938_fsc.xml emd_15938_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_15938.png emd_15938.png | 84 KB | ||
| Masks |  emd_15938_msk_1.map emd_15938_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Others |  emd_15938_additional_1.map.gz emd_15938_additional_1.map.gz emd_15938_half_map_1.map.gz emd_15938_half_map_1.map.gz emd_15938_half_map_2.map.gz emd_15938_half_map_2.map.gz | 287.4 MB 279.6 MB 279.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15938 http://ftp.pdbj.org/pub/emdb/structures/EMD-15938 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15938 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15938 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15938.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15938.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Globally sharpened consensus map generated using RELION Postprocess | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||
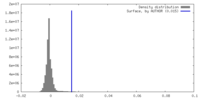
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15938_msk_1.map emd_15938_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
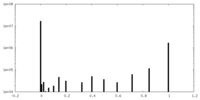
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened and unfiltered consensus map generated using RELION...
| File | emd_15938_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened and unfiltered consensus map generated using RELION Postprocess | ||||||||||||
| Projections & Slices |
| ||||||||||||
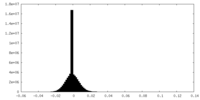
| Density Histograms |
-Half map: Half map 2 generated using RELION Postprocess (unsharpened...
| File | emd_15938_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 generated using RELION Postprocess (unsharpened and unfiltered) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 generated using RELION Postprocess (unsharpened...
| File | emd_15938_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 generated using RELION Postprocess (unsharpened and unfiltered) | ||||||||||||
| Projections & Slices |
| ||||||||||||
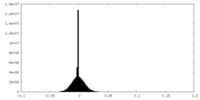
| Density Histograms |
- Sample components
Sample components
-Entire : Mitochondrial respiratory complex I
| Entire | Name: Mitochondrial respiratory complex I |
|---|---|
| Components |
|
-Supramolecule #1: Mitochondrial respiratory complex I
| Supramolecule | Name: Mitochondrial respiratory complex I / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#43 Details: Native purification of mitochondrial complex I from Drosophila melanogaster (fruit fly) W1118. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. Details: Following glow discharge for 90 s at 20 mA, the grid was treated for 7 days in an anaerobic glovebox in ethanol containing 5 mM 11-mercaptoundecyl hexaethyleneglycol, washed three times in ...Details: Following glow discharge for 90 s at 20 mA, the grid was treated for 7 days in an anaerobic glovebox in ethanol containing 5 mM 11-mercaptoundecyl hexaethyleneglycol, washed three times in ethanol and dried prior to blotting | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 10 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3082 / Average exposure time: 10.0 sec. / Average electron dose: 41.88 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)