[English] 日本語
 Yorodumi
Yorodumi- EMDB-15893: CryoEM Structure of Extended eEF1A bound to the Ribosome in the C... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of Extended eEF1A bound to the Ribosome in the Classical Pre State | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | translation / protein biogenesis / elongation factor / ribosome | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of lipid kinase activity / cytoplasmic side of lysosomal membrane / Eukaryotic Translation Elongation / eukaryotic translation elongation factor 1 complex / regulation of chaperone-mediated autophagy / translation factor activity, RNA binding / translational elongation / translation elongation factor activity / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / positive regulation of apoptotic process ...positive regulation of lipid kinase activity / cytoplasmic side of lysosomal membrane / Eukaryotic Translation Elongation / eukaryotic translation elongation factor 1 complex / regulation of chaperone-mediated autophagy / translation factor activity, RNA binding / translational elongation / translation elongation factor activity / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / positive regulation of apoptotic process / translation / GTPase activity / synapse / protein kinase binding / endoplasmic reticulum membrane / GTP binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||
 Authors Authors | Gemmer M / Fedry JMM / Forster FG | ||||||||||||
| Funding support | European Union,  Netherlands, 3 items Netherlands, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Visualization of translation and protein biogenesis at the ER membrane. Authors: Max Gemmer / Marten L Chaillet / Joyce van Loenhout / Rodrigo Cuevas Arenas / Dimitrios Vismpas / Mariska Gröllers-Mulderij / Fujiet A Koh / Pascal Albanese / Richard A Scheltema / Stuart C ...Authors: Max Gemmer / Marten L Chaillet / Joyce van Loenhout / Rodrigo Cuevas Arenas / Dimitrios Vismpas / Mariska Gröllers-Mulderij / Fujiet A Koh / Pascal Albanese / Richard A Scheltema / Stuart C Howes / Abhay Kotecha / Juliette Fedry / Friedrich Förster /  Abstract: The dynamic ribosome-translocon complex, which resides at the endoplasmic reticulum (ER) membrane, produces a major fraction of the human proteome. It governs the synthesis, translocation, membrane ...The dynamic ribosome-translocon complex, which resides at the endoplasmic reticulum (ER) membrane, produces a major fraction of the human proteome. It governs the synthesis, translocation, membrane insertion, N-glycosylation, folding and disulfide-bond formation of nascent proteins. Although individual components of this machinery have been studied at high resolution in isolation, insights into their interplay in the native membrane remain limited. Here we use cryo-electron tomography, extensive classification and molecular modelling to capture snapshots of mRNA translation and protein maturation at the ER membrane at molecular resolution. We identify a highly abundant classical pre-translocation intermediate with eukaryotic elongation factor 1a (eEF1a) in an extended conformation, suggesting that eEF1a may remain associated with the ribosome after GTP hydrolysis during proofreading. At the ER membrane, distinct polysomes bind to different ER translocons specialized in the synthesis of proteins with signal peptides or multipass transmembrane proteins with the translocon-associated protein complex (TRAP) present in both. The near-complete atomic model of the most abundant ER translocon variant comprising the protein-conducting channel SEC61, TRAP and the oligosaccharyltransferase complex A (OSTA) reveals specific interactions of TRAP with other translocon components. We observe stoichiometric and sub-stoichiometric cofactors associated with OSTA, which are likely to include protein isomerases. In sum, we visualize ER-bound polysomes with their coordinated downstream machinery. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15893.map.gz emd_15893.map.gz | 268.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15893-v30.xml emd-15893-v30.xml emd-15893.xml emd-15893.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15893.png emd_15893.png | 135.7 KB | ||
| Filedesc metadata |  emd-15893.cif.gz emd-15893.cif.gz | 7.5 KB | ||
| Others |  emd_15893_half_map_1.map.gz emd_15893_half_map_1.map.gz emd_15893_half_map_2.map.gz emd_15893_half_map_2.map.gz | 383.6 MB 383.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15893 http://ftp.pdbj.org/pub/emdb/structures/EMD-15893 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15893 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15893 | HTTPS FTP |
-Related structure data
| Related structure data |  8b6zMC  8b6lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15893.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15893.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.999 Å | ||||||||||||||||||||||||||||||||||||
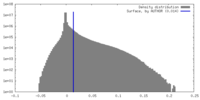
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15893_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
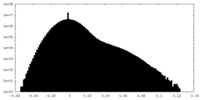
| Projections & Slices |
| ||||||||||||
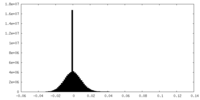
| Density Histograms |
-Half map: #1
| File | emd_15893_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
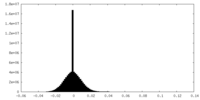
| Density Histograms |
- Sample components
Sample components
-Entire : Ribosome in the Classical Pre+ state
| Entire | Name: Ribosome in the Classical Pre+ state |
|---|---|
| Components |
|
-Supramolecule #1: Ribosome in the Classical Pre+ state
| Supramolecule | Name: Ribosome in the Classical Pre+ state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Elongation factor 1-alpha 2
| Macromolecule | Name: Elongation factor 1-alpha 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.545102 KDa |
| Sequence | String: MGKEKTHINI VVIGHVDSGK STTTGHLIYK CGGIDKRTIE KFEKEAAEMG KGSFKYAWVL DKLKAERERG ITIDISLWKF ETTKYYITI IDAPGHRDFI KNMITGTSQA DCAVLIVAAG VGEFEAGISK NGQTREHALL AYTLGVKQLI VGVNKMDSTE P AYSEKRYD ...String: MGKEKTHINI VVIGHVDSGK STTTGHLIYK CGGIDKRTIE KFEKEAAEMG KGSFKYAWVL DKLKAERERG ITIDISLWKF ETTKYYITI IDAPGHRDFI KNMITGTSQA DCAVLIVAAG VGEFEAGISK NGQTREHALL AYTLGVKQLI VGVNKMDSTE P AYSEKRYD EIVKEVSAYI KKIGYNPATV PFVPISGWHG DNMLEPSPNM PWFKGWKVER KEGNASGVSL LEALDTILPP TR PTDKPLR LPLQDVYKIG GIGTVPVGRV ETGILRPGMV VTFAPVNITT EVKSVEMHHE ALSEALPGDN VGFNVKNVSV KDI RRGNVC GDSKSDPPQE AAQFTSQVII LNHPGQISAG YSPVIDCHTA HIACKFAELK EKIDRRSGKK LEDNPKSLKS GDAA IVEMV PGKPMCVESF SQYPPLGRFA VRDMRQTVAV GVIKNVEKKS GGAGKVTKSA QKAQKAGK UniProtKB: Elongation factor 1-alpha 2 |
-Macromolecule #2: 28S ribosomal RNA
| Macromolecule | Name: 28S ribosomal RNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.640222125 MDa |
| Sequence | String: CGCGACCUCA GAUCAGACGU GGCGACCCGC UGAAUUUAAG CAUAUUAGUC AGCGGAGGAG AAGAAACUAA CCAGGAUUCC CUCAGUAAC GGCGAGUGAA CAGGGAAGAG CCCAGCGCCG AAUCCCCGCC CCGCGGCGGG GCGCGGGACA UGUGGCGUAC G GAAGACCC ...String: CGCGACCUCA GAUCAGACGU GGCGACCCGC UGAAUUUAAG CAUAUUAGUC AGCGGAGGAG AAGAAACUAA CCAGGAUUCC CUCAGUAAC GGCGAGUGAA CAGGGAAGAG CCCAGCGCCG AAUCCCCGCC CCGCGGCGGG GCGCGGGACA UGUGGCGUAC G GAAGACCC GCUCCCCGGC GCCGCUCGUG GGGGGCCCAA GUCCUUCUGA UCGAGGCCCA GCCCGUGGAC GGUGUGAGGC CG GUAGCGG CCCCCGGCGC GCCGGGCCCG GGUCUUCCCG GAGUCGGGUU GCUUGGGAAU GCAGCCCAAA GCGGGUGGUA AAC UCCAUC UAAGGCUAAA UACCGGCACG AGACCGAUAG UCAACAAGUA CCGUAAGGGA AAGUUGAAAA GAACUUUGAA GAGA GAGUU CAAGAGGGCG UGAAACCGUU AAGAGGUAAA CGGGUGGGGU CCGCGCAGUC CGCCCGGAGG AUUCAACCCG GCGGC GGGU CCGGCCGUGU CGGCGGCCCG GCGGAUCUUU CCCGCCCCCC GUUCCUCCCG ACCCCUCCAC CCGCCCUCCC UUCCCC CGC CGCCCCUCCU CCUCCUCCCC GGAGGGGGCG GGCUCCGGCG GGUGCGGGGG UGGGCGGGCG GGGCCGGGGG UGGGGUC GG CGGGGGACCG UCCCCCGACC GGCGACCGGC CGCCGCCGGG CGCAUUUCCA CCGCGGCGGU GCGCCGCGAC CGGCUCCG G GACGGCUGGG AAGGCCCGGC GGGGAAGGUG GCUCGGGGGG CCCCGUCCGU CCGUCCGUCC GUCCUCCUCC UCCCCCGUC UCCGCCCCCC GGCCCCGCGU CCUCCCUCGG GAGGGCGCGC GGGUCGGGGC GGCGGCGGCG GCGGCGGUGG CGGCGGCGGC GGCGGCGGC GGGACCGAAA CCCCCCCCGA GUGUUACAGC CCCCCCGGCA GCAGCACUCG CCGAAUCCCG GGGCCGAGGG A GCGAGACC CGUCGCCGCG CUCUCCCCCC UCCCGGCGCC CACCCCCGCG GGGAAUCCCC CGCGAGGGGG GUCUCCCCCG CG GGGGCGC GCCGGCGUCU CCUCGUGGGG GGGCCGGGCC ACCCCUCCCA CGGCGCGACC GCUCUCCCAC CCCUCCUCCC CGC GCCCCC GCCCCGGCGA CGGGGGGGGU GCCGCGCGCG GGUCGGGGGG CGGGGCGGAC UGUCCCCAGU GCGCCCCGGG CGGG UCGCG CCGUCGGGCC CGGGGGAGGU UCUCUCGGGG CCACGCGCGC GUCCCCCGAA GAGGGGGACG GCGGAGCGAG CGCAC GGGG UCGGCGGCGA CGUCGGCUAC CCACCCGACC CGUCUUGAAA CACGGACCAA GGAGUCUAAC ACGUGCGCGA GUCGGG GGC UCGCACGAAA GCCGCCGUGG CGCAAUGAAG GUGAAGGCCG GCGCGCUCGC CGGCCGAGGU GGGAUCCCGA GGCCUCU CC AGUCCGCCGA GGGCGCACCA CCGGCCCGUC UCGCCCGCCG CGCCGGGGAG GUGGAGCACG AGCGCACGUG UUAGGACC C GAAAGAUGGU GAACUAUGCC UGGGCAGGGC GAAGCCAGAG GAAACUCUGG UGGAGGUCCG UAGCGGUCCU GACGUGCAA AUCGGUCGUC CGACCUGGGU AUAGGGGCGA AAGACUAAUC GAACCAUCUA GUAGCUGGUU CCCUCCGAAG UUUCCCUCAG GAUAGCUGG CGCUCUCGCA GACCCGACGC ACCCCCGCCA CGCAGUUUUA UCCGGUAAAG CGAAUGAUUA GAGGUCUUGG G GCCGAAAC GAUCUCAACC UAUUCUCAAA CUUUAAAUGG GUAAGAAGCC CGGCUCGCUG GCGUGGAGCC GGGCGUGGAA UG CGAGUGC CUAGUGGGCC ACUUUUGGUA AGCAGAACUG GCGCUGCGGG AUGAACCGAA CGCCGGGUUA AGGCGCCCGA UGC CGACGC UCAUCAGACC CCAGAAAAGG UGUUGGUUGA UAUAGACAGC AGGACGGUGG CCAUGGAAGU CGGAAUCCGC UAAG GAGUG UGUAACAACU CACCUGCCGA AUCAACUAGC CCUGAAAAUG GAUGGCGCUG GAGCGUCGGG CCCAUACCCG GCCGU CGCC GGCAGUCGAG AGUGGACGGG AGCGGCGGGG GCGGCGCGCG CGCGCGCGCG UGUGGUGUGC GUCGGAGGGC GGCGGC GGC GGCGGCGGCG GGGGUGUGGG GUCCUUCCCC CGCCCCCCCC CCCACGCCUC CUCCCCUCCU CCCGCCCACG CCCCGCU CC CCGCCCCCGG AGCCCCGCGG ACGCUACGCC GCGACGAGUA GGAGGGCCGC UGCGGUGAGC CUUGAAGCCU AGGGCGCG G GCCCGGGUGG AGCCGCCGCA GGUGCAGAUC UUGGUGGUAG UAGCAAAUAU UCAAACGAGA ACUUUGAAGG CCGAAGUGG AGAAGGGUUC CAUGUGAACA GCAGUUGAAC AUGGGUCAGU CGGUCCUGAG AGAUGGGCGA GCGCCGUUCC GAAGGGACGG GCGAUGGCC UCCGUUGCCC UCGGCCGAUC GAAAGGGAGU CGGGUUCAGA UCCCCGAAUC CGGAGUGGCG GAGAUGGGCG C CGCGAGGC GUCCAGUGCG GUAACGCGAC CGAUCCCGGA GAAGCCGGCG GGAGCCCCGG GGAGAGUUCU CUUUUCUUUG UG AAGGGCA GGGCGCCCUG GAAUGGGUUC GCCCCGAGAG AGGGGCCCGU GCCUUGGAAA GCGUCGCGGU UCCGGCGGCG UCC GGUGAG CUCUCGCUGG CCCUUGAAAA UCCGGGGGAG AGGGUGUAAA UCUCGCGCCG GGCCGUACCC AUAUCCGCAG CAGG UCUCC AAGGUGAACA GCCUCUGGCA UGUUGGAACA AUGUAGGUAA GGGAAGUCGG CAAGCCGGAU CCGUAACUUC GGGAU AAGG AUUGGCUCUA AGGGCUGGGU CGGUCGGGCU GGGGCGCGAA GCGGGGCUGG GCGCGCGCCG CGGCUGGACG AGGCGC CGC CGCCCCCCCC ACGCCCGGGG CACCCCCCUC GCGGCCCUCC CCCGCCCCAC CCCGCGCGCG CCGCUCGCUC CCUCCCC GC CCCGCGCCCU CUCUCUCUCU CUCUCCCCCG CUCCCCGUCC UCCCCCCUCC CCGGGGGAGC GCCGCGUGGG GGCGGCGG C GGGGGGAGAA GGGUCGGGGC GGCAGGGGCC GGCGGCGGCC CGCCGCGGGG CCCCGGCGGC GGGGGCACGG UCCCCCGCG AGGGGGGCCC GGGCACCCGG GGGGCCGGCG GCGGCGGCGA CUCUGGACGC GAGCCGGGCC CUUCCCGUGG AUCGCCCCAG CUGCGGCGG GCGUCGCGGC CGCCCCCGGG GAGCCCGGCG GGCGCCGGCG CGCCCCCCCC CCCACCCCAC GUCUCGUCGC G CGCGCGUC CGCUGGGGGC GGGGAGCGGU CGGGCGGCGG CGGUCGGCGG GCGGCGGGGC GGGGCGGUUC GUCCCCCCGC CC UACCCCC CCGGCCCCGU CCGCCCCCCG UUCCCCCCUC CUCCUCGGCG CGCGGCGGCG GCGGCGGCAG GCGGCGGAGG GGC CGCGGG CCGGUCCCCC CCGCCGGGUC CGCCCCCGGG GCCGCGGUUC CGCGCGGCGC CUCGCCUCGG CCGGCGCCUA GCAG CCGAC UUAGAACUGG UGCGGACCAG GGGAAUCCGA CUGUUUAAUU AAAACAAAGC AUCGCGAAGG CCCGCGGCGG GUGUU GACG CGAUGUGAUU UCUGCCCAGU GCUCUGAAUG UCAAAGUGAA GAAAUUCAAU GAAGCGCGGG UAAACGGCGG GAGUAA CUA UGACUCUCUU AAGGUAGCCA AAUGCCUCGU CAUCUAAUUA GUGACGCGCA UGAAUGGAUG AACGAGAUUC CCACUGU CC CUACCUACUA UCCAGCGAAA CCACAGCCAA GGGAACGGGC UUGGCGGAAU CAGCGGGGAA AGAAGACCCU GUUGAGCU U GACUCUAGUC UGGCACGGUG AAGAGACAUG AGAGGUGUAG AAUAAGUGGG AGGCCCCCGG CGCCCCCCCG GUGUCCCCG CGAGGGGCCC GGGGCGGGGU CCGCCGGCCC UGCGGGCCGC CGGUGAAAUA CCACUACUCU GAUCGUUUUU UCACUGACCC GGUGAGGCG GGGGGGCGAG CCCCGAGGGG CUCUCGCUUC UGGCGCCAAG CGCCCGGCCG CGCGCCGGCC GGGCGCGACC C GCUCCGGG GACAGUGCCA GGUGGGGAGU UUGACUGGGG CGGUACACCU GUCAAACGGU AACGCAGGUG UCCUAAGGCG AG CUCAGGG AGGACAGAAA CCUCCCGUGG AGCAGAAGGG CAAAAGCUCG CUUGAUCUUG AUUUUCAGUA CGAAUACAGA CCG UGAAAG CGGGGCCUCA CGAUCCUUCU GACCUUUUGG GUUUUAAGCA GGAGGUGUCA GAAAAGUUAC CACAGGGAUA ACUG GCUUG UGGCGGCCAA GCGUUCAUAG CGACGUCGCU UUUUGAUCCU UCGAUGUCGG CUCUUCCUAU CAUUGUGAAG CAGAA UUCA CCAAGCGUUG GAUUGUUCAC CCACUAAUAG GGAACGUGAG CUGGGUUUAG ACCGUCGUGA GACAGGUUAG UUUUAC CCU ACUGAUGAUG UGUUGUUGCC AUGGUAAUCC UGCUCAGUAC GAGAGGAACC GCAGGUUCAG ACAUUUGGUG UAUGUGC UU GGCUGAGGAG CCAAUGGGGC GAAGCUACCA UCUGUGGGAU UAUGACUGAA CGCCUCUAAG UCAGAAUCCC GCCCAGGC G GAACGAUACG GCAGCGCCGC GGAGCCUCGG UUGGCCUCGG AUAGCCGGUC CCCCGCCUGU CCCCGCCGGC GGGCCGCCC CCCCCCUCCA CGCGCCCCGC GCGCGCGGGA GGGCGCGUGC CCCGCCGCGC GCCGGGACCG GGGUCCGGUG CGGAGUGCCC UUCGUCCUG GGAAACGGGG CGCGGCCGGA GAGGCGGCCG CCCCCUCGCC CGUCACGCAC CGCACGUUCG UGGGGAACCU G GCGCUAAA CCAUUCGUAG ACGACCUGCU UCUGGGUCGG GGUUUCGUAC GUAGCAGAGC AGCUCCCUCG CUGCGAUCUA UU GAAAGUC AGCCCUCGAC ACAAGGGUUU GUC GENBANK: GENBANK: AL592188.60 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.2 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 19046 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final 3D classification | Software - Name: RELION |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)