[English] 日本語
 Yorodumi
Yorodumi- EMDB-15693: Structure of Bacillus pseudofirmus Mrp antiporter complex, monome... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Bacillus pseudofirmus Mrp antiporter complex, monomer, processed at original pixel size (0.837 A/pix) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Alkalihalobacillus pseudofirmus (bacteria) Alkalihalobacillus pseudofirmus (bacteria) | |||||||||
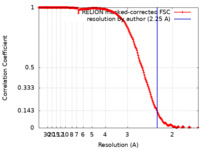
| Method | single particle reconstruction / cryo EM / Resolution: 2.25 Å | |||||||||
 Authors Authors | Lee Y | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Ion transfer mechanisms in Mrp-type antiporters from high resolution cryoEM and molecular dynamics simulations. Authors: Yongchan Lee / Outi Haapanen / Anton Altmeyer / Werner Kühlbrandt / Vivek Sharma / Volker Zickermann /    Abstract: Multiple resistance and pH adaptation (Mrp) cation/proton antiporters are essential for growth of a variety of halophilic and alkaliphilic bacteria under stress conditions. Mrp-type antiporters are ...Multiple resistance and pH adaptation (Mrp) cation/proton antiporters are essential for growth of a variety of halophilic and alkaliphilic bacteria under stress conditions. Mrp-type antiporters are closely related to the membrane domain of respiratory complex I. We determined the structure of the Mrp antiporter from Bacillus pseudofirmus by electron cryo-microscopy at 2.2 Å resolution. The structure resolves more than 99% of the sidechains of the seven membrane subunits MrpA to MrpG plus 360 water molecules, including ~70 in putative ion translocation pathways. Molecular dynamics simulations based on the high-resolution structure revealed details of the antiport mechanism. We find that switching the position of a histidine residue between three hydrated pathways in the MrpA subunit is critical for proton transfer that drives gated trans-membrane sodium translocation. Several lines of evidence indicate that the same histidine-switch mechanism operates in respiratory complex I. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15693.map.gz emd_15693.map.gz | 27.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15693-v30.xml emd-15693-v30.xml emd-15693.xml emd-15693.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15693_fsc.xml emd_15693_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_15693.png emd_15693.png | 97.3 KB | ||
| Masks |  emd_15693_msk_1.map emd_15693_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_15693_half_map_1.map.gz emd_15693_half_map_1.map.gz emd_15693_half_map_2.map.gz emd_15693_half_map_2.map.gz | 414 MB 414 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15693 http://ftp.pdbj.org/pub/emdb/structures/EMD-15693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15693 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15693.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15693.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15693_msk_1.map emd_15693_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15693_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15693_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MrpABCDEFG complex
| Entire | Name: MrpABCDEFG complex |
|---|---|
| Components |
|
-Supramolecule #1: MrpABCDEFG complex
| Supramolecule | Name: MrpABCDEFG complex / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Alkalihalobacillus pseudofirmus (bacteria) Alkalihalobacillus pseudofirmus (bacteria) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)