[English] 日本語
 Yorodumi
Yorodumi- EMDB-15643: Native Chemotaxis Core Signalling Complex from E. coli, Focused a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Native Chemotaxis Core Signalling Complex from E. coli, Focused alignment on ligand binding domain | |||||||||||||||||||||
 Map data Map data | Native Chemotaxis Core Signalling Complex in E. coli, focused alignment on the ligand binding domain, cubic map, unfiltered, unmasked | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Chemotaxis Core Signalling Complex from E. coli / Chemotaxis / Signal Transduction / Chemoreceptor / Core signaling complex / MEMBRANE PROTEIN | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 12.5 Å | |||||||||||||||||||||
 Authors Authors | Qin Z / Zhang P | |||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, European Union, 6 items United States, European Union, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: mBio / Year: 2023 Journal: mBio / Year: 2023Title: Structure of the native chemotaxis core signaling unit from phage E-protein lysed cells. Authors: C Keith Cassidy / Zhuan Qin / Thomas Frosio / Khoosheh Gosink / Zhengyi Yang / Mark S P Sansom / Phillip J Stansfeld / John S Parkinson / Peijun Zhang /   Abstract: Bacterial chemotaxis is a ubiquitous behavior that enables cell movement toward or away from specific chemicals. It serves as an important model for understanding cell sensory signal transduction and ...Bacterial chemotaxis is a ubiquitous behavior that enables cell movement toward or away from specific chemicals. It serves as an important model for understanding cell sensory signal transduction and motility. Characterization of the molecular mechanisms underlying chemotaxis is of fundamental interest and requires a high-resolution structural picture of the sensing machinery, the chemosensory array. In this study, we combine cryo-electron tomography and molecular simulation to present the complete structure of the core signaling unit, the basic building block of chemosensory arrays, from . Our results provide new insight into previously poorly-resolved regions of the complex and offer a structural basis for designing new experiments to test mechanistic hypotheses. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15643.map.gz emd_15643.map.gz | 25 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15643-v30.xml emd-15643-v30.xml emd-15643.xml emd-15643.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15643.png emd_15643.png | 82.7 KB | ||
| Filedesc metadata |  emd-15643.cif.gz emd-15643.cif.gz | 4.2 KB | ||
| Others |  emd_15643_half_map_1.map.gz emd_15643_half_map_1.map.gz emd_15643_half_map_2.map.gz emd_15643_half_map_2.map.gz | 25 MB 25 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15643 http://ftp.pdbj.org/pub/emdb/structures/EMD-15643 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15643 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15643 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15643.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15643.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Native Chemotaxis Core Signalling Complex in E. coli, focused alignment on the ligand binding domain, cubic map, unfiltered, unmasked | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.745 Å | ||||||||||||||||||||||||||||||||||||
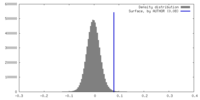
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map. Native Chemotaxis Core Signalling Complex in...
| File | emd_15643_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map. Native Chemotaxis Core Signalling Complex in E. coli, focused alignment on the ligand binding domain, cubic map, unfiltered, unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
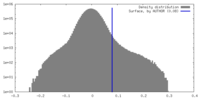
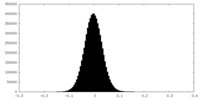
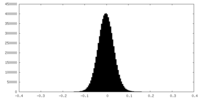
| Density Histograms |
-Half map: half map. Native Chemotaxis Core Signalling Complex in...
| File | emd_15643_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map. Native Chemotaxis Core Signalling Complex in E. coli, focused alignment on the ligand binding domain, cubic map, unfiltered, unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
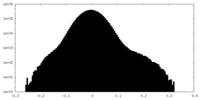
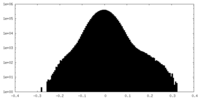
| Density Histograms |
- Sample components
Sample components
-Entire : Native Chemotaxis Core Signalling Complex
| Entire | Name: Native Chemotaxis Core Signalling Complex |
|---|---|
| Components |
|
-Supramolecule #1: Native Chemotaxis Core Signalling Complex
| Supramolecule | Name: Native Chemotaxis Core Signalling Complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 12.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number subtomograms used: 5100 |
|---|---|
| Extraction | Number tomograms: 33 / Number images used: 20000 |
| Final angle assignment | Type: OTHER / Software - Name: PROTOMO (ver. 0.9) |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)