+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Consensus map of dynein-dynactin-BICDR on microtubules | ||||||||||||
 Map data Map data | Consensus map of Dynein-Dynactin-BICDR on microtubules | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | ||||||||||||
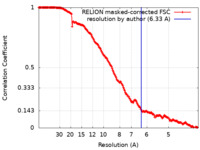
| Method | single particle reconstruction / cryo EM / Resolution: 6.33 Å | ||||||||||||
 Authors Authors | Chaaban S / Carter AP | ||||||||||||
| Funding support |  United Kingdom, European Union, 3 items United Kingdom, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structure of dynein-dynactin on microtubules shows tandem adaptor binding. Authors: Sami Chaaban / Andrew P Carter /  Abstract: Cytoplasmic dynein is a microtubule motor that is activated by its cofactor dynactin and a coiled-coil cargo adaptor. Up to two dynein dimers can be recruited per dynactin, and interactions between ...Cytoplasmic dynein is a microtubule motor that is activated by its cofactor dynactin and a coiled-coil cargo adaptor. Up to two dynein dimers can be recruited per dynactin, and interactions between them affect their combined motile behaviour. Different coiled-coil adaptors are linked to different cargos, and some share motifs known to contact sites on dynein and dynactin. There is limited structural information on how the resulting complex interacts with microtubules and how adaptors are recruited. Here we develop a cryo-electron microscopy processing pipeline to solve the high-resolution structure of dynein-dynactin and the adaptor BICDR1 bound to microtubules. This reveals the asymmetric interactions between neighbouring dynein motor domains and how they relate to motile behaviour. We found that two adaptors occupy the complex. Both adaptors make similar interactions with the dyneins but diverge in their contacts with each other and dynactin. Our structure has implications for the stability and stoichiometry of motor recruitment by cargos. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15396.map.gz emd_15396.map.gz | 202.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15396-v30.xml emd-15396-v30.xml emd-15396.xml emd-15396.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15396_fsc.xml emd_15396_fsc.xml | 17.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15396.png emd_15396.png | 75 KB | ||
| Masks |  emd_15396_msk_1.map emd_15396_msk_1.map | 216 MB |  Mask map Mask map | |
| Others |  emd_15396_half_map_1.map.gz emd_15396_half_map_1.map.gz emd_15396_half_map_2.map.gz emd_15396_half_map_2.map.gz | 193.9 MB 193.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15396 http://ftp.pdbj.org/pub/emdb/structures/EMD-15396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15396 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15396.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15396.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map of Dynein-Dynactin-BICDR on microtubules | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.489 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15396_msk_1.map emd_15396_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 of Dynein-Dynactin-BICDR on microtubules
| File | emd_15396_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of Dynein-Dynactin-BICDR on microtubules | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 of Dynein-Dynactin-BICDR on microtubules
| File | emd_15396_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of Dynein-Dynactin-BICDR on microtubules | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of dynein, dynactin, and BICDR1 bound to microtubules
| Entire | Name: Complex of dynein, dynactin, and BICDR1 bound to microtubules |
|---|---|
| Components |
|
-Supramolecule #1: Complex of dynein, dynactin, and BICDR1 bound to microtubules
| Supramolecule | Name: Complex of dynein, dynactin, and BICDR1 bound to microtubules type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Molecular weight | Theoretical: 4 MDa |
-Supramolecule #2: Dynein, cytoplasmic 1
| Supramolecule | Name: Dynein, cytoplasmic 1 / type: organelle_or_cellular_component / ID: 2 / Parent: 1 / Macromolecule list: #13-#16 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.4 MDa |
| Recombinant expression | Organism:  |
-Supramolecule #3: Dynactin
| Supramolecule | Name: Dynactin / type: organelle_or_cellular_component / ID: 3 / Parent: 1 / Macromolecule list: #1-#10 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.1 MDa |
-Supramolecule #4: BICDR1
| Supramolecule | Name: BICDR1 / type: organelle_or_cellular_component / ID: 4 / Parent: 1 / Macromolecule list: #11 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 130 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 Component:
| |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV / Details: 20 second incubation. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 14 / Number real images: 88715 / Average exposure time: 3.0 sec. / Average electron dose: 53.0 e/Å2 Details: Images were collected in movie-mode and fractionated into 53 movie frames |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)