[English] 日本語
 Yorodumi
Yorodumi- EMDB-15322: Complex III2 from Yarrowia lipolytica,antimycin A bound, int-position -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex III2 from Yarrowia lipolytica,antimycin A bound, int-position | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | oxidoreductase / electron transport chain / inhibitor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial processing peptidase complex / mitochondrial processing peptidase / matrix side of mitochondrial inner membrane / : / mitochondrial respiratory chain complex III assembly / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / mitochondrial crista ...mitochondrial processing peptidase complex / mitochondrial processing peptidase / matrix side of mitochondrial inner membrane / : / mitochondrial respiratory chain complex III assembly / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / mitochondrial crista / nuclear periphery / metalloendopeptidase activity / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / oxidoreductase activity / mitochondrial inner membrane / heme binding / mitochondrion / proteolysis / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) | |||||||||
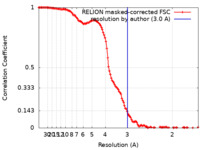
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Wieferig JP / Kuhlbrandt W | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2023 Journal: IUCrJ / Year: 2023Title: Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III by cryo-EM. Authors: Jan Philip Wieferig / Werner Kühlbrandt /  Abstract: Movement of the Rieske domain of the iron-sulfur protein is essential for intramolecular electron transfer within complex III (CIII) of the respiratory chain as it bridges a gap in the cofactor chain ...Movement of the Rieske domain of the iron-sulfur protein is essential for intramolecular electron transfer within complex III (CIII) of the respiratory chain as it bridges a gap in the cofactor chain towards the electron acceptor cytochrome c. We present cryo-EM structures of CIII from Yarrowia lipolytica at resolutions up to 2.0 Å under different conditions, with different redox states of the cofactors of the high-potential chain. All possible permutations of three primary positions were observed, indicating that the two halves of the dimeric complex act independently. Addition of the substrate analogue decylubiquinone to CIII with a reduced high-potential chain increased the occupancy of the Q site. The extent of Rieske domain interactions through hydrogen bonds to the cytochrome b and cytochrome c subunits varied depending on the redox state and substrate. In the absence of quinols, the reduced Rieske domain interacted more closely with cytochrome b and cytochrome c than in the oxidized state. Upon addition of the inhibitor antimycin A, the heterogeneity of the cd-helix and ef-loop increased, which may be indicative of a long-range effect on the Rieske domain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15322.map.gz emd_15322.map.gz | 161.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15322-v30.xml emd-15322-v30.xml emd-15322.xml emd-15322.xml | 27.4 KB 27.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15322_fsc.xml emd_15322_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
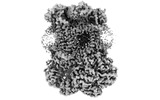
| Images |  emd_15322.png emd_15322.png | 65.4 KB | ||
| Filedesc metadata |  emd-15322.cif.gz emd-15322.cif.gz | 7.7 KB | ||
| Others |  emd_15322_half_map_1.map.gz emd_15322_half_map_1.map.gz emd_15322_half_map_2.map.gz emd_15322_half_map_2.map.gz | 140.6 MB 140.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15322 http://ftp.pdbj.org/pub/emdb/structures/EMD-15322 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15322 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15322 | HTTPS FTP |
-Related structure data
| Related structure data |  8abiMC  8ab6C  8ab7C  8ab8C  8ab9C  8abaC  8abbC  8abeC  8abfC  8abgC  8abhC  8abjC  8abkC  8ablC  8abmC  8ac3C  8ac4C  8ac5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15322.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15322.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15322_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15322_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Complex III2, with antmiycin A, int-position
+Supramolecule #1: Complex III2, with antmiycin A, int-position
+Macromolecule #1: Cytochrome b
+Macromolecule #2: Cytochrome b-c1 complex subunit Rieske, mitochondrial
+Macromolecule #3: Cytochrome b-c1 complex subunit 7
+Macromolecule #4: YALI0F24673p
+Macromolecule #5: YALI0A14806p
+Macromolecule #6: Cytochrome b-c1 complex subunit 2, mitochondrial
+Macromolecule #7: YALI0A17468p
+Macromolecule #8: Cytochrome b-c1 complex subunit 8
+Macromolecule #9: Complex III subunit 9
+Macromolecule #10: YALI0C12210p
+Macromolecule #11: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #12: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #13: PHOSPHATIDYLETHANOLAMINE
+Macromolecule #14: CARDIOLIPIN
+Macromolecule #15: [(2R,3S,6S,7R,8R)-3-[(3-formamido-2-oxidanyl-phenyl)carbonylamino...
+Macromolecule #16: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #17: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #18: 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE
+Macromolecule #19: HEME C
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)