[English] 日本語
 Yorodumi
Yorodumi- EMDB-1510: Structure of full-length Epac2 in complex with cyclic-AMP and Rap. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1510 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of full-length Epac2 in complex with cyclic-AMP and Rap. | |||||||||
 Map data Map data | This is a map of the Epac2-cAMP-Rap1B complex. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GEF / GTPases / Epac / Rap / cAMP | |||||||||
| Function / homology | : / intracellular anatomical structure / Ras guanine-nucleotide exchange factors catalytic domain / :  Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Arias-Palomo E / Rehmann H / Hadders M / Schwede F / Bos JL / Llorca O | |||||||||
 Citation Citation |  Journal: Nature / Year: 2008 Journal: Nature / Year: 2008Title: Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Authors: Holger Rehmann / Ernesto Arias-Palomo / Michael A Hadders / Frank Schwede / Oscar Llorca / Johannes L Bos /  Abstract: Epac proteins are activated by binding of the second messenger cAMP and then act as guanine nucleotide exchange factors for Rap proteins. The Epac proteins are involved in the regulation of cell ...Epac proteins are activated by binding of the second messenger cAMP and then act as guanine nucleotide exchange factors for Rap proteins. The Epac proteins are involved in the regulation of cell adhesion and insulin secretion. Here we have determined the structure of Epac2 in complex with a cAMP analogue (Sp-cAMPS) and RAP1B by X-ray crystallography and single particle electron microscopy. The structure represents the cAMP activated state of the Epac2 protein with the RAP1B protein trapped in the course of the exchange reaction. Comparison with the inactive conformation reveals that cAMP binding causes conformational changes that allow the cyclic nucleotide binding domain to swing from a position blocking the Rap binding site towards a docking site at the Ras exchange motif domain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1510.map.gz emd_1510.map.gz | 404.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1510-v30.xml emd-1510-v30.xml emd-1510.xml emd-1510.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1510.gif 1510.gif | 51 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1510 http://ftp.pdbj.org/pub/emdb/structures/EMD-1510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1510 | HTTPS FTP |
-Validation report
| Summary document |  emd_1510_validation.pdf.gz emd_1510_validation.pdf.gz | 200.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1510_full_validation.pdf.gz emd_1510_full_validation.pdf.gz | 199.2 KB | Display | |
| Data in XML |  emd_1510_validation.xml.gz emd_1510_validation.xml.gz | 4.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1510 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1510 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1510 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1510 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1510.map.gz / Format: CCP4 / Size: 422.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1510.map.gz / Format: CCP4 / Size: 422.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of the Epac2-cAMP-Rap1B complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
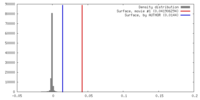
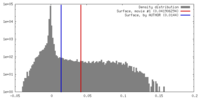
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Epac2-cAMP-Rap1B complex
| Entire | Name: Epac2-cAMP-Rap1B complex |
|---|---|
| Components |
|
-Supramolecule #1000: Epac2-cAMP-Rap1B complex
| Supramolecule | Name: Epac2-cAMP-Rap1B complex / type: sample / ID: 1000 Oligomeric state: One monomer of Epac2 binds one monomer of Rap1B in presence of cAMP Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 135 KDa |
-Macromolecule #1: Rap1B
| Macromolecule | Name: Rap1B / type: protein_or_peptide / ID: 1 / Name.synonym: Rap1B / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 18 KDa |
| Sequence | GO: intracellular anatomical structure / InterPro: INTERPRO: IPR003577 |
-Macromolecule #2: Epac2
| Macromolecule | Name: Epac2 / type: protein_or_peptide / ID: 2 / Name.synonym: Epac2 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 115 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: GO: 0019933 InterPro: Ras guanine-nucleotide exchange factors catalytic domain |
-Macromolecule #3: Adenosine cyclic monophosphate
| Macromolecule | Name: Adenosine cyclic monophosphate / type: ligand / ID: 3 / Name.synonym: cAMP / Details: cAMP is the Epac2 activator. / Recombinant expression: No |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 329.21 Da |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: 50mM Tris-HCL, 100mM NaCl, 10mM CaCl2, 500 uM cAMP, 5% glycerol. |
|---|---|
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein floated on 2% w/v uranyl formate for 60 seconds |
| Grid | Details: 400 mesh Copper/Palladium grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1230 |
|---|---|
| Details | Microscope JEOL 1230 |
| Date | Jul 19, 2007 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 10.5 µm / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.9 mm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
- Image processing
Image processing
| CTF correction | Details: Phase correction at the micrograph level |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN,Xmipp / Number images used: 3442 |
| Final two d classification | Number classes: 233 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)