[English] 日本語
 Yorodumi
Yorodumi- EMDB-1504: Structural basis for the regulated protease and chaperone functio... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1504 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis for the regulated protease and chaperone function of DegP | |||||||||
 Map data Map data | negative stain EM structure of the DegP24-OMP complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | protease-chaperone / electron microscopy / single particle analysis | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidase Do / response to temperature stimulus / : / protein quality control for misfolded or incompletely synthesized proteins / serine-type peptidase activity / peptidase activity / outer membrane-bounded periplasmic space / protein folding / response to heat / response to oxidative stress ...peptidase Do / response to temperature stimulus / : / protein quality control for misfolded or incompletely synthesized proteins / serine-type peptidase activity / peptidase activity / outer membrane-bounded periplasmic space / protein folding / response to heat / response to oxidative stress / periplasmic space / serine-type endopeptidase activity / proteolysis / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Krojer T / Sawa J / Schaefer E / Saibil HR / Ehrmann M / Clausen T | |||||||||
 Citation Citation |  Journal: Nature / Year: 2008 Journal: Nature / Year: 2008Title: Structural basis for the regulated protease and chaperone function of DegP. Authors: Tobias Krojer / Justyna Sawa / Eva Schäfer / Helen R Saibil / Michael Ehrmann / Tim Clausen /  Abstract: All organisms have to monitor the folding state of cellular proteins precisely. The heat-shock protein DegP is a protein quality control factor in the bacterial envelope that is involved in ...All organisms have to monitor the folding state of cellular proteins precisely. The heat-shock protein DegP is a protein quality control factor in the bacterial envelope that is involved in eliminating misfolded proteins and in the biogenesis of outer-membrane proteins. Here we describe the molecular mechanisms underlying the regulated protease and chaperone function of DegP from Escherichia coli. We show that binding of misfolded proteins transforms hexameric DegP into large, catalytically active 12-meric and 24-meric multimers. A structural analysis of these particles revealed that DegP represents a protein packaging device whose central compartment is adaptable to the size and concentration of substrate. Moreover, the inner cavity serves antagonistic functions. Whereas the encapsulation of folded protomers of outer-membrane proteins is protective and might allow safe transit through the periplasm, misfolded proteins are eliminated in the molecular reaction chamber. Oligomer reassembly and concomitant activation on substrate binding may also be critical in regulating other HtrA proteases implicated in protein-folding diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1504.map.gz emd_1504.map.gz | 7.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1504-v30.xml emd-1504-v30.xml emd-1504.xml emd-1504.xml | 6.9 KB 6.9 KB | Display Display |  EMDB header EMDB header |
| Images |  1504.gif 1504.gif | 77.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1504 http://ftp.pdbj.org/pub/emdb/structures/EMD-1504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1504 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1504.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1504.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | negative stain EM structure of the DegP24-OMP complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.44 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
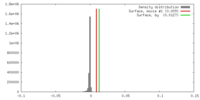
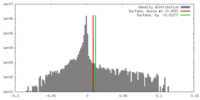
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : proteolytically inactive DegP24mer with bound Omp protein
| Entire | Name: proteolytically inactive DegP24mer with bound Omp protein |
|---|---|
| Components |
|
-Supramolecule #1000: proteolytically inactive DegP24mer with bound Omp protein
| Supramolecule | Name: proteolytically inactive DegP24mer with bound Omp protein type: sample / ID: 1000 / Details: monodisperse sample / Oligomeric state: 24mer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 1.13 MDa |
-Macromolecule #1: protease
| Macromolecule | Name: protease / type: protein_or_peptide / ID: 1 / Name.synonym: DegP Details: proteolytically inactive DegP24mer with bound Omp protein Number of copies: 1 / Oligomeric state: 24 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.0064 mg/mL |
|---|---|
| Staining | Type: NEGATIVE Details: negatively stained with 2% (w/v) uranyl acetate on glow discharged, carbon-coated grids |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Sample stage | Specimen holder: single tilt / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: O (octahedral) / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF Details: octahedral symmetry was used for the reconstruction |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)