+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human Apoferritin obtained from ssDNA coated grid | |||||||||
 Map data Map data | Final map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human 80S ribosome / ssDNA covered grid / ssDNA / cryoEM / RIBOSOME | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Hrebik D / Plevka P | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2022 Journal: Acta Crystallogr D Struct Biol / Year: 2022Title: Polyelectrolyte coating of cryo-EM grids improves lateral distribution and prevents aggregation of macromolecules. Authors: Dominik Hrebík / Mária Gondová / Lucie Valentová / Tibor Füzik / Antonín Přidal / Jiří Nováček / Pavel Plevka /  Abstract: Cryo-electron microscopy (cryo-EM) is one of the primary methods used to determine the structures of macromolecules and their complexes. With the increased availability of cryo-electron microscopes, ...Cryo-electron microscopy (cryo-EM) is one of the primary methods used to determine the structures of macromolecules and their complexes. With the increased availability of cryo-electron microscopes, the preparation of high-quality samples has become a bottleneck in the cryo-EM structure-determination pipeline. Macromolecules can be damaged during the purification or preparation of vitrified samples for cryo-EM, making them prone to binding to the grid support, to aggregation or to the adoption of preferential orientations at the air-water interface. Here, it is shown that coating cryo-EM grids with a negatively charged polyelectrolyte, such as single-stranded DNA, before applying the sample reduces the aggregation of macromolecules and improves their distribution. The single-stranded DNA-coated grids enabled the determination of high-resolution structures from samples that aggregated on conventional grids. The polyelectrolyte coating reduces the diffusion of macromolecules and thus may limit the negative effects of the contact of macromolecules with the grid support and blotting paper, as well as of the shear forces on macromolecules during grid blotting. Coating grids with polyelectrolytes can readily be employed in any laboratory dealing with cryo-EM sample preparation, since it is fast, simple, inexpensive and does not require specialized equipment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14704.map.gz emd_14704.map.gz | 94.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14704-v30.xml emd-14704-v30.xml emd-14704.xml emd-14704.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
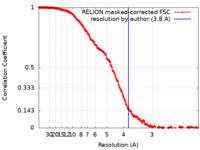
| FSC (resolution estimation) |  emd_14704_fsc.xml emd_14704_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14704.png emd_14704.png | 81.6 KB | ||
| Filedesc metadata |  emd-14704.cif.gz emd-14704.cif.gz | 6.9 KB | ||
| Others |  emd_14704_half_map_1.map.gz emd_14704_half_map_1.map.gz emd_14704_half_map_2.map.gz emd_14704_half_map_2.map.gz | 94.6 MB 94.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14704 http://ftp.pdbj.org/pub/emdb/structures/EMD-14704 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14704 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14704 | HTTPS FTP |
-Related structure data
| Related structure data |  7zfwMC  7ze1C  7zg7C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14704.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14704.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
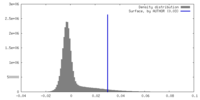
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_14704_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
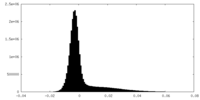
| Density Histograms |
-Half map: Half Map 2
| File | emd_14704_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
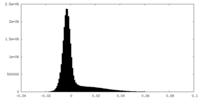
| Density Histograms |
- Sample components
Sample components
-Entire : human 80S ribosome
| Entire | Name: human 80S ribosome |
|---|---|
| Components |
|
-Supramolecule #1: human 80S ribosome
| Supramolecule | Name: human 80S ribosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.3 MDa |
-Macromolecule #1: RNA (615-MER)
| Macromolecule | Name: RNA (615-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.222873375 MDa |
| Sequence | String: CGCGACCUCA GAUCAGACGU GGCGACCCGC UGAAUUUAAG CAUAUUAGUC AGCGGAGGAG AAGAAACUAA CCAGGAUUCC CUCAGUAAC GGCGAGUGAA CAGGGAAGAG CCCAGCGCCG AAUCCCCGCC CCGCGGCGGG GCGCGGGACA UGUGGCGUAC G GAAGACCC ...String: CGCGACCUCA GAUCAGACGU GGCGACCCGC UGAAUUUAAG CAUAUUAGUC AGCGGAGGAG AAGAAACUAA CCAGGAUUCC CUCAGUAAC GGCGAGUGAA CAGGGAAGAG CCCAGCGCCG AAUCCCCGCC CCGCGGCGGG GCGCGGGACA UGUGGCGUAC G GAAGACCC GCUCCCCGGC GCCGCUCGUG GGGGGCCCAA GUCCUUCUGA UCGAGGCCCA GCCCGUGGAC GGUGUGAGGC CG GUAGCGG CCCCCGGCGC GCCGGGCCCG GGUCUUCCCG GAGUCGGGUU GCUUGGGAAU GCAGCCCAAA GCGGGUGGUA AAC UCCAUC UAAGGCUAAA UACCGGCACG AGACCGAUAG UCAACAAGUA CCGUAA(OMG)GGA AAGUUGAAAA GAACUUUGAA G(A2M)GAGAGUU CAAGAGGGCG UGAAACCGUU AAGAGGUAAA CGGGUGGGGU CCGCGCAGUC CGCCCGGAGG AUUCAAC CC GGCGGCGGGU CCGGCCGUGU CGGCGGCCCG GCGGAUCUUU CCCGCGCGGG GGACCGUCCC CCGACCGGCG ACCGGCCG C CGCCGGGCGC AUUUCCACCG CGGCGGUGCG CCGCGACCGG CUCCGGGACG GCU(2MG)GGGAAG GCCCGGCGGG GAAGG UGGC UCGGGGGCCC CCGAGUGUUA CAGCCCCCCC GGCAGCAGCA CUCGCCGAAU CCCGGGGCCG AGGGAGCGAG ACCCGU CGC CGCGCUCUCC CCCCUCCCGG CGCGCCGGGG GGGGCCGGGC CACCCCUCCC ACGGCGCGAC CGCUCGGGGC GGACUGU CC CCAGUGCGCC CCGGGCGGGU CGCGCCGUCG GGCCCGGGGG AGGCCACGCG CGCGUCCCCC GAAGAGGGGG ACGGCGGA G CGAGCGCACG GGGUCGGCGG CGACGUCGGC UACCCACCCG ACCCGUCUUG AAACACGGAC CAAGGAGUCU AACACGUGC GCGAGUCGGG GGCUCGCACG AAAGCCGCCG UGGCGCAAUG AAGGUGAAGG CCGGCGCGCU CGCCGGCCGA GGUGGGAUCC CGAGGCCUC UCCAGUCCGC CGAGGGCGCA CCACCGGCCC GUCUCGCCCG CCGCGCCGGG GAGGUGGAGC ACGAGCGCAC G UGUUAGGA CCCGAAAGAU GGUGAACUAU GCCUGGGCAG GGCGAAGCCA GAGGAAACUC UGGUGGAGGU CCGUAGCGGU CC UGACGUG CAAAUCGGUC GUCCGACCUG GGUAUAGGGG CGAAAGACUA AUCGAACCAU CUAGUAGCUG GUUCCCUCCG AAG UUUCCC UCAGGAUAGC UGGCGCUCUC GCAGACCCGA CGCCCGCCAC GCAGUUUUAU CCGGUAAAGC GAAUGAUUAG AGGU CUUGG GGCCGAAACG AUCUCAACCU AUUCUCAAAC UUUAAAUGGG UAAGAAGCCC GGCUCGCUGG CGUGGAGCCG GGCGU GGAA UGCGAGUGCC UAGUGGGCCA CUUUUGGUAA GCAGAACUGG CGCUGCGGGA UGAACCGAAC GCCGGGUUAA GGCGCC CGA UGCCGACGCU CAUCAGACCC CAGAAAAGGU GUUGGUUGAU AUAGACAGCA GGACGGUGGC CAUGGAAGUC GGAAUCC GC UAAGGAGUGU GUAACAACUC ACCUGCCGAA UCAACUAGCC CUGAAAAUGG AUGGCGCUGG AGCGUCGGGC CCAUACCC G GCCGUCGCCG GCAGUCGAGA GUGGACGGGA GCGGCGGGCC GGAGCCCCGC GGACGCUACG CCGCGACGAG UAGGAGGGC CGCUGCGGUG AGCCUUGAAG CCUAGGGCGC GGGCCCGGGU GGAGCCGCCG CAGGUGCAGA UCUUGGUGGU AGUAGCAAAU AUUCAAACG AGAACUUUGA AGGCCGAAGU GGAGAAGGGU UCCAUGUGAA CAGCAGUUGA ACAUGGGUCA GUCGGUCCUG A GAGAUGGG CGAGCGCCGU UCCGAAGGGA CGGGCGAUGG CCUCCGUUGC CCUCGGCCGA UCGAAAGGGA GUCGGGUUCA GA UCCCCGA AUCCGGAGUG GCGGAGAUGG GCGCCGCGAG GCGUCCAGUG CGGUAACGCG ACCGAUCCCG GAGAAGCCGG CGG GAGCCC CGGGGAGAGU UCUCUUUUCU UUGUGAAGGG CAGGGCGCCC UGGAAUGGGU UCGCCCCGAG AGAGGGGCCC GUGC CUUGG AAAGCGUCGC GGUUCCGGCG GCGUCCGGUG AGCUCUCGCU GGCCCUUGAA AAUCCGGGGG AGAGGGUGUA AAUCU CGCG CCGGGCCGUA CCCAUAUCCG CAGCAGGUCU CCAAGGUGAA CAGCCUCUGG CAUGUUGGAA CAAUGUAGGU AAGGGA AGU CGGCAAGCCG GAUCCGUAAC UUCGGGAUAA GGAUUGGCUC UAAGGGCUGG GUCGGUCGCG GCCGGCGCCU AGCAGCC GA CUUAGAACUG GUGCGGACCA GGGGAAUCCG ACUGUUUAAU UAAAACAAAG CAUCGCGAAG GCCCGCGGCG GGUGUUGA C GCGAUGUGAU UUCUGCCCAG UGCUCUGAAU GUCAAAGUGA AGAAAUUCAA UGAAGCGCGG GUAAACGGCG GGAGUAACU AUGACUCUCU UAAGGUAGCC AAAUGCCUCG UCAUCUAAUU AGUGACGCGC AUGAAUGGAU GAACGAGAUU CCCACUGUCC CUACCUACU AUCCAGCGAA ACCACAGCCA AGGGAACGGG CUUGGCGGAA UCAGCGGGGA AAGAAGACCC UGUUGAGCUU G ACUCUAGU CUGGCACGGU GAAGAGACAU GAGAGGUGUA GAAUAAGUGG GAGGCCCCCG GCGCCCCCCC GGUGUCCCCG CG AGGGGCC CGGGGCGGGG UCCGCCGGCC CUGCGGGCCG CCGGUGAAAU ACCACUACUC UGAUCGUUUU UUCACUGACC CGG UGAGGC GGGGGGGCGA GCCCCGAGGG GCUCUCGCUU CUGGCGCCAA GCGCCCGGCC GCGCGCCGGC CGGGCGCGAC CCGC UCCGG GGACAGUGCC AGGUGGGGAG UUUGACUGGG GCGGUACACC UGUCAAACGG UAACGCAGGU GUCCUAAGGC GAGCU CAGG GAGGACAGAA ACCUCCCGUG GAGCAGAAGG GCAAAAGCUC GCUUGAUCUU GAUUUUCAGU ACGAAUACAG ACCGUG AAA GCGGGGCCUC ACGAUCCUUC UGACCUUUUG GGUUUUAAGC AGGAGGUGUC AGAAAAGUUA CCACAGGGAU AACUGGC UU GUGGCGGCCA AGCGUUCAUA GCGACGUCGC UUUUUGAUCC UUCGAUGUCG GCUCUUCCUA UCAUUGUGAA GCAGAAUU C ACCAAGCGUU GGAUUGUUCA CCCACUAAUA GGGAACGUGA GCUGGGUUUA GACCGUCGUG AGACAGGUUA GUUUUACCC UACUGAUGAU GUGUUGUUGC CAUGGUAAUC CUGCUCAGUA CGAGAGGAAC CGCAGGUUCA GACAUUUGGU GUAUGUGCUU GGCUGAGGA GCCAAUGGGG CGAAGCUACC AUCUGUGGGA UUAUGACUGA ACGCCUCUAA GUCAGAAUCC CGCCCAGGCG G AACGAUAC GGCAGCGCCG CGGAGCCUCG GUUGGCCUCG GAUAGCCGGU CCCCCGCCGG GGUCCGGUGC GGAGUGCCCU UC GUCCUGG GAAACGGGGC GCGGCCGGAG AGGCGGCCGC CCCCUCGCCC GUCACGCACC GCACGUUCGU GGGGAACCUG GCG CUAAAC CAUUCGUAGA CGACCUGCUU CUGGGUCGGG GUUUCGUACG UAGCAGAGCA GCUCCCUCGC UGCGAUCUAU UGAA AGUCA GCCCUCGACA CAAGGGUUUG U |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 5 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: NITROGEN | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: 3 s blot time,30 s waiting time, ssDNA covered grid. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 4527 / Average exposure time: 1.0 sec. / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)