[English] 日本語
 Yorodumi
Yorodumi- EMDB-14623: Subtomogram averaging of Rubisco from Cyanobium carboxysome (inne... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram averaging of Rubisco from Cyanobium carboxysome (innermost layer) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Cyanobium sp. PCC 7001 (bacteria) Cyanobium sp. PCC 7001 (bacteria) | ||||||||||||
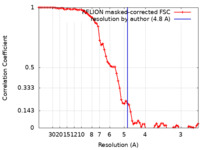
| Method | subtomogram averaging / cryo EM / Resolution: 4.8 Å | ||||||||||||
 Authors Authors | Ni T / Seaton-Burn W / Zhu Y / Zhang P | ||||||||||||
| Funding support |  United Kingdom, European Union, 3 items United Kingdom, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure and assembly of cargo Rubisco in two native α-carboxysomes. Authors: Tao Ni / Yaqi Sun / Will Burn / Monsour M J Al-Hazeem / Yanan Zhu / Xiulian Yu / Lu-Ning Liu / Peijun Zhang /   Abstract: Carboxysomes are a family of bacterial microcompartments in cyanobacteria and chemoautotrophs. They encapsulate Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) and carbonic anhydrase ...Carboxysomes are a family of bacterial microcompartments in cyanobacteria and chemoautotrophs. They encapsulate Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) and carbonic anhydrase catalyzing carbon fixation inside a proteinaceous shell. How Rubisco complexes pack within the carboxysomes is unknown. Using cryo-electron tomography, we determine the distinct 3D organization of Rubisco inside two distant α-carboxysomes from a marine α-cyanobacterium Cyanobium sp. PCC 7001 where Rubiscos are organized in three concentric layers, and from a chemoautotrophic bacterium Halothiobacillus neapolitanus where they form intertwining spirals. We further resolve the structures of native Rubisco as well as its higher-order assembly at near-atomic resolutions by subtomogram averaging. The structures surprisingly reveal that the authentic intrinsically disordered linker protein CsoS2 interacts with Rubiscos in native carboxysomes but functions distinctively in the two α-carboxysomes. In contrast to the uniform Rubisco-CsoS2 association in the Cyanobium α-carboxysome, CsoS2 binds only to the Rubiscos close to the shell in the Halo α-carboxysome. Our findings provide critical knowledge of the assembly principles of α-carboxysomes, which may aid in the rational design and repurposing of carboxysome structures for new functions. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14623.map.gz emd_14623.map.gz | 32.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14623-v30.xml emd-14623-v30.xml emd-14623.xml emd-14623.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14623_fsc.xml emd_14623_fsc.xml | 7.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14623.png emd_14623.png | 130.7 KB | ||
| Masks |  emd_14623_msk_1.map emd_14623_msk_1.map | 34.3 MB |  Mask map Mask map | |
| Others |  emd_14623_half_map_1.map.gz emd_14623_half_map_1.map.gz emd_14623_half_map_2.map.gz emd_14623_half_map_2.map.gz | 20.2 MB 20.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14623 http://ftp.pdbj.org/pub/emdb/structures/EMD-14623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14623 | HTTPS FTP |
-Validation report
| Summary document |  emd_14623_validation.pdf.gz emd_14623_validation.pdf.gz | 871.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14623_full_validation.pdf.gz emd_14623_full_validation.pdf.gz | 871 KB | Display | |
| Data in XML |  emd_14623_validation.xml.gz emd_14623_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_14623_validation.cif.gz emd_14623_validation.cif.gz | 18.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14623 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14623 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14623 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14623 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14623.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14623.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||
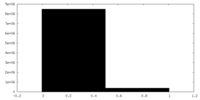
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14623_msk_1.map emd_14623_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
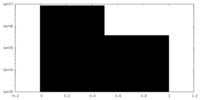
| Projections & Slices |
| ||||||||||||
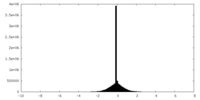
| Density Histograms |
-Half map: emClarity half map 1 from cisTEM reconstruction
| File | emd_14623_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | emClarity half map 1 from cisTEM reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
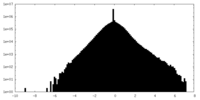
| Density Histograms |
-Half map: emClarity half map 2 from cisTEM reconstruction
| File | emd_14623_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | emClarity half map 2 from cisTEM reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rubisco from Cyanobium carboxysome (innermost layer)
| Entire | Name: Rubisco from Cyanobium carboxysome (innermost layer) |
|---|---|
| Components |
|
-Supramolecule #1: Rubisco from Cyanobium carboxysome (innermost layer)
| Supramolecule | Name: Rubisco from Cyanobium carboxysome (innermost layer) / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Cyanobium sp. PCC 7001 (bacteria) Cyanobium sp. PCC 7001 (bacteria) |
-Macromolecule #1: Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) large s...
| Macromolecule | Name: Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cyanobium sp. PCC 7001 (bacteria) Cyanobium sp. PCC 7001 (bacteria) |
| Sequence | String: MSKKYDAGVK EYRDTYWTPD YVPLDTDLLA CFKCTGQEGV PKEEVAAAVA AESSTGTWST VWSELLVDL DFYKGRCYRI EDVPGDKEAF YAFIAYPLDL FEEGSVTNVL TSLVGNVFGF K ALRHLRLE DIRFPMAFIK TCPGPPNGIC VERDRMNKYG RPLLGCTIKP ...String: MSKKYDAGVK EYRDTYWTPD YVPLDTDLLA CFKCTGQEGV PKEEVAAAVA AESSTGTWST VWSELLVDL DFYKGRCYRI EDVPGDKEAF YAFIAYPLDL FEEGSVTNVL TSLVGNVFGF K ALRHLRLE DIRFPMAFIK TCPGPPNGIC VERDRMNKYG RPLLGCTIKP KLGLSGKNYG RV VYECLRG GLDFTKDDEN INSQPFQRWQ NRFEFVAEAV ALAQQETGEK KGHYLNCTAA TPE EMYERA EFAKELGQPI IMHDYITGGF TANTGLSKWC RKNGMLLHIH RAMHAVIDRH PKHG IHFRV LAKCLRLSGG DQLHTGTVVG KLEGDRQTTL GFIDQLRESF IPEDRSRGNF FDQDW GSMP GVFAVASGGI HVWHMPALVA IFGDDSVLQF GGGTHGHPWG SAAGAAANRV ALEACV KAR NAGREIEKES RDILMEAAKH SPELAIALET WKEIKFEFDT VDKLDVQ |
-Macromolecule #2: Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) small s...
| Macromolecule | Name: Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cyanobium sp. PCC 7001 (bacteria) Cyanobium sp. PCC 7001 (bacteria) |
| Sequence | String: MPFKSTVGDY QTVATLETFG FLPPMTQDEI YDQIAYIIAQ GWSPLIEHVH PSRSMATYWS YWKLPFFGE KDLGVIVSEL EACHRAYPDH HVRLVGYDAY TQSQGACFVV FEGR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 3.65 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)