+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14369 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of USP9X, local refinement of monomer | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | USP9X / Deubiquitinase / ubiquitin / Structural Genomics / Structural Genomics Consortium / SGC / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytosolic ciliogenesis / K11-linked deubiquitinase activity / protein import into peroxisome matrix, receptor recycling / protein deubiquitination involved in ubiquitin-dependent protein catabolic process / co-SMAD binding / female gamete generation / monoubiquitinated protein deubiquitination / positive regulation of TORC2 signaling / deubiquitinase activity / DNA alkylation repair ...cytosolic ciliogenesis / K11-linked deubiquitinase activity / protein import into peroxisome matrix, receptor recycling / protein deubiquitination involved in ubiquitin-dependent protein catabolic process / co-SMAD binding / female gamete generation / monoubiquitinated protein deubiquitination / positive regulation of TORC2 signaling / deubiquitinase activity / DNA alkylation repair / axon extension / K48-linked deubiquitinase activity / protein K63-linked deubiquitination / RHOV GTPase cycle / K63-linked deubiquitinase activity / RHOU GTPase cycle / molecular sequestering activity / cilium assembly / protein deubiquitination / BMP signaling pathway / positive regulation of protein binding / cysteine-type peptidase activity / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / transforming growth factor beta receptor signaling pathway / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / chromosome segregation / Peroxisomal protein import / Downregulation of SMAD2/3:SMAD4 transcriptional activity / regulation of circadian rhythm / regulation of protein stability / neuron migration / intracellular protein localization / cell migration / rhythmic process / growth cone / amyloid fibril formation / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / Ub-specific processing proteases / protein stabilization / cilium / protein ubiquitination / Amyloid fiber formation / cell division / cysteine-type endopeptidase activity / centrosome / negative regulation of transcription by RNA polymerase II / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
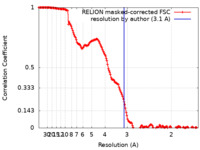
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Deme JC / Halabelian L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of USP9X Authors: Halabelian L / Deme JC / Lea SM / Arrowsmith CH / Structural Genomics Consortium (SGC) | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
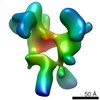
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14369.map.gz emd_14369.map.gz | 324.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14369-v30.xml emd-14369-v30.xml emd-14369.xml emd-14369.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14369_fsc.xml emd_14369_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14369.png emd_14369.png | 131.2 KB | ||
| Masks |  emd_14369_msk_1.map emd_14369_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14369.cif.gz emd-14369.cif.gz | 7.4 KB | ||
| Others |  emd_14369_additional_1.map.gz emd_14369_additional_1.map.gz emd_14369_half_map_1.map.gz emd_14369_half_map_1.map.gz emd_14369_half_map_2.map.gz emd_14369_half_map_2.map.gz | 171.8 MB 318.2 MB 318.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14369 http://ftp.pdbj.org/pub/emdb/structures/EMD-14369 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14369 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14369 | HTTPS FTP |
-Validation report
| Summary document |  emd_14369_validation.pdf.gz emd_14369_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14369_full_validation.pdf.gz emd_14369_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_14369_validation.xml.gz emd_14369_validation.xml.gz | 23.6 KB | Display | |
| Data in CIF |  emd_14369_validation.cif.gz emd_14369_validation.cif.gz | 31.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14369 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14369 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14369 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14369 | HTTPS FTP |
-Related structure data
| Related structure data |  7yxyMC  7yxxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14369.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14369.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
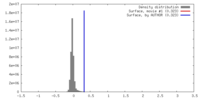
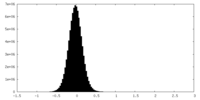
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_14369_msk_1.map emd_14369_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
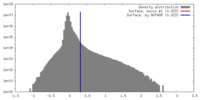
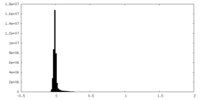
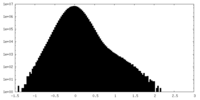
| Density Histograms |
-Additional map: unsharpened map
| File | emd_14369_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
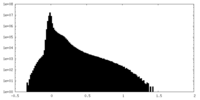
| Density Histograms |
-Half map: half map 1
| File | emd_14369_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
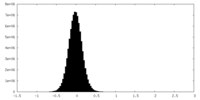
| Density Histograms |
-Half map: half map 2
| File | emd_14369_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
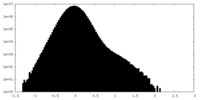
| Density Histograms |
- Sample components
Sample components
-Entire : USP9X monomer
| Entire | Name: USP9X monomer |
|---|---|
| Components |
|
-Supramolecule #1: USP9X monomer
| Supramolecule | Name: USP9X monomer / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Probable ubiquitin carboxyl-terminal hydrolase FAF-X
| Macromolecule | Name: Probable ubiquitin carboxyl-terminal hydrolase FAF-X / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ubiquitinyl hydrolase 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 293.878844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHSSG RENLYFQGMT ATTRGSPVGG NDNQGQAPDG QSQPPLQQNQ TSSPDSSNEN SPATPPDEQG QGDAPPQLED EEPAFPHTD LAKLDDMINR PRWVVPVLPK GELEVLLEAA IDLSKKGLDV KSEACQRFFR DGLTISFTKI LTDEAVSGWK F EIHRCIIN ...String: MHHHHHHSSG RENLYFQGMT ATTRGSPVGG NDNQGQAPDG QSQPPLQQNQ TSSPDSSNEN SPATPPDEQG QGDAPPQLED EEPAFPHTD LAKLDDMINR PRWVVPVLPK GELEVLLEAA IDLSKKGLDV KSEACQRFFR DGLTISFTKI LTDEAVSGWK F EIHRCIIN NTHRLVELCV AKLSQDWFPL LELLAMALNP HCKFHIYNGT RPCESVSSSV QLPEDELFAR SPDPRSPKGW LV DLLNKFG TLNGFQILHD RFINGSALNV QIIAALIKPF GQCYEFLTLH TVKKYFLPII EMVPQFLENL TDEELKKEAK NEA KNDALS MIIKSLKNLA SRVPGQEETV KNLEIFRLKM ILRLLQISSF NGKMNALNEV NKVISSVSYY THRHGNPEEE EWLT AERMA EWIQQNNILS IVLRDSLHQP QYVEKLEKIL RFVIKEKALT LQDLDNIWAA QAGKHEAIVK NVHDLLAKLA WDFSP EQLD HLFDCFKASW TNASKKQREK LLELIRRLAE DDKDGVMAHK VLNLLWNLAH SDDVPVDIMD LALSAHIKIL DYSCSQ DRD TQKIQWIDRF IEELRTNDKW VIPALKQIRE ICSLFGEAPQ NLSQTQRSPH VFYRHDLINQ LQHNHALVTL VAENLAT YM ESMRLYARDH EDYDPQTVRL GSRYSHVQEV QERLNFLRFL LKDGQLWLCA PQAKQIWKCL AENAVYLCDR EACFKWYS K LMGDEPDLDP DINKDFFESN VLQLDPSLLT ENGMKCFERF FKAVNCREGK LVAKRRAYMM DDLELIGLDY LWRVVIQSN DDIASRAIDL LKEIYTNLGP RLQVNQVVIH EDFIQSCFDR LKASYDTLCV LDGDKDSVNC ARQEAVRMVR VLTVLREYIN ECDSDYHEE RTILPMSRAF RGKHLSFVVR FPNQGRQVDD LEVWSHTNDT IGSVRRCILN RIKANVAHTK IELFVGGELI D PADDRKLI GQLNLKDKSL ITAKLTQISS NMPSSPDSSS DSSTGSPGNH GNHYSDGPNP EVESCLPGVI MSLHPRYISF LW QVADLGS SLNMPPLRDG ARVLMKLMPP DSTTIEKLRA ICLDHAKLGE SSLSPSLDSL FFGPSASQVL YLTEVVYALL MPA GAPLAD DSSDFQFHFL KSGGLPLVLS MLTRNNFLPN ADMETRRGAY LNALKIAKLL LTAIGYGHVR AVAEACQPGV EGVN PMTQI NQVTHDQAVV LQSALQSIPN PSSECMLRNV SVRLAQQISD EASRYMPDIC VIRAIQKIIW ASGCGSLQLV FSPNE EITK IYEKTNAGNE PDLEDEQVCC EALEVMTLCF ALIPTALDAL SKEKAWQTFI IDLLLHCHSK TVRQVAQEQF FLMCTR CCM GHRPLLFFIT LLFTVLGSTA RERAKHSGDY FTLLRHLLNY AYNSNINVPN AEVLLNNEID WLKRIRDDVK RTGETGI EE TILEGHLGVT KELLAFQTSE KKFHIGCEKG GANLIKELID DFIFPASNVY LQYMRNGELP AEQAIPVCGS PPTINAGF E LLVALAVGCV RNLKQIVDSL TEMYYIGTAI TTCEALTEWE YLPPVGPRPP KGFVGLKNAG ATCYMNSVIQ QLYMIPSIR NGILAIEGTG SDVDDDMSGD EKQDNESNVD PRDDVFGYPQ QFEDKPALSK TEDRKEYNIG VLRHLQVIFG HLAASRLQYY VPRGFWKQF RLWGEPVNLR EQHDALEFFN SLVDSLDEAL KALGHPAMLS KVLGGSFADQ KICQGCPHRY ECEESFTTLN V DIRNHQNL LDSLEQYVKG DLLEGANAYH CEKCNKKVDT VKRLLIKKLP PVLAIQLKRF DYDWERECAI KFNDYFEFPR EL DMEPYTV AGVAKLEGDN VNPESQLIQQ SEQSESETAG STKYRLVGVL VHSGQASGGH YYSYIIQRNG GDGERNRWYK FDD GDVTEC KMDDDEEMKN QCFGGEYMGE VFDHMMKRMS YRRQKRWWNA YILFYERMDT IDQDDELIRY ISELAITTRP HQII MPSAI ERSVRKQNVQ FMHNRMQYSM EYFQFMKKLL TCNGVYLNPP PGQDHLLPEA EEITMISIQL AARFLFTTGF HTKKV VRGS ASDWYDALCI LLRHSKNVRF WFAHNVLFNV SNRFSEYLLE CPSAEVRGAF AKLIVFIAHF SLQDGPCPSP FASPGP SSQ AYDNLSLSDH LLRAVLNLLR REVSEHGRHL QQYFNLFVMY ANLGVAEKTQ LLKLSVPATF MLVSLDEGPG PPIKYQY AE LGKLYSVVSQ LIRCCNVSSR MQSSINGNPP LPNPFGDPNL SQPIMPIQQN VADILFVRTS YVKKIIEDCS NSEETVKL L RFCCWENPQF SSTVLSELLW QVAYSYTYEL RPYLDLLLQI LLIEDSWQTH RIHNALKGIP DDRDGLFDTI QRSKNHYQK RAYQCIKCMV ALFSNCPVAY QILQGNGDLK RKWTWAVEWL GDELERRPYT GNPQYTYNNW SPPVQSNETS NGYFLERSHS ARMTLAKAC ELCPEEEPDD QDAPDEHESP PPEDAPLYPH SPGSQYQQNN HVHGQPYTGP AAHHMNNPQR TGQRAQENYE G SEEVSPPQ TKDQDYKDDD K UniProtKB: Ubiquitin carboxyl-terminal hydrolase 9X |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 56.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)