[English] 日本語
 Yorodumi
Yorodumi- EMDB-1424: The structure of the prokaryotic cyclic nucleotide-modulated pota... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1424 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the prokaryotic cyclic nucleotide-modulated potassium channel MloK1 at 16 A resolution. | |||||||||
 Map data Map data | This is the 3D map of MloK1 in 17-A resolution determined by NS-TEM. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Mesorhizobium loti (bacteria) / synthetic construct (others) Mesorhizobium loti (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 16.3 Å | |||||||||
 Authors Authors | Chiu P-L / Pagel M / Evans J / Chou H-T / Gipson B | |||||||||
 Citation Citation |  Journal: Structure / Year: 2007 Journal: Structure / Year: 2007Title: The structure of the prokaryotic cyclic nucleotide-modulated potassium channel MloK1 at 16 A resolution. Authors: Po-Lin Chiu / Matthew D Pagel / James Evans / Hui-Ting Chou / Xiangyan Zeng / Bryant Gipson / Henning Stahlberg / Crina M Nimigean /  Abstract: The gating ring of cyclic nucleotide-modulated channels is proposed to be either a two-fold symmetric dimer of dimers or a four-fold symmetric tetramer based on high-resolution structure data of ...The gating ring of cyclic nucleotide-modulated channels is proposed to be either a two-fold symmetric dimer of dimers or a four-fold symmetric tetramer based on high-resolution structure data of soluble cyclic nucleotide-binding domains and functional data on intact channels. We addressed this controversy by obtaining structural data on an intact, full-length, cyclic nucleotide-modulated potassium channel, MloK1, from Mesorhizobium loti, which also features a putative voltage-sensor. We present here the 3D single-particle structure by transmission electron microscopy and the projection map of membrane-reconstituted 2D crystals of MloK1 in the presence of cAMP. Our data show a four-fold symmetric arrangement of the CNBDs, separated by discrete gaps. A homology model for full-length MloK1 suggests a vertical orientation for the CNBDs. The 2D crystal packing in the membrane-embedded state is compatible with the S1-S4 domains in the vertical "up" state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
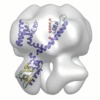
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1424.map.gz emd_1424.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1424-v30.xml emd-1424-v30.xml emd-1424.xml emd-1424.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1424.gif 1424.gif | 67.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1424 http://ftp.pdbj.org/pub/emdb/structures/EMD-1424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1424 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1424.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1424.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the 3D map of MloK1 in 17-A resolution determined by NS-TEM. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
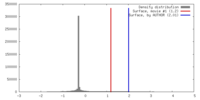
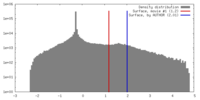
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Full-length prokaryotic potassium channel MloK1
| Entire | Name: Full-length prokaryotic potassium channel MloK1 |
|---|---|
| Components |
|
-Supramolecule #1000: Full-length prokaryotic potassium channel MloK1
| Supramolecule | Name: Full-length prokaryotic potassium channel MloK1 / type: sample / ID: 1000 / Oligomeric state: One tetramer with ligand binding / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 155.2 KDa / Theoretical: 152.5 KDa / Method: Calculation by the protein sequence |
-Macromolecule #1: Prokaryotic potassium channel
| Macromolecule | Name: Prokaryotic potassium channel / type: protein_or_peptide / ID: 1 / Name.synonym: MloK1 / Details: Each subunit of the MloK1 / Number of copies: 1 / Oligomeric state: Tetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Mesorhizobium loti (bacteria) / Strain: MloK1 / synonym: Soil bacteria / Tissue: Prokaryote / Organelle: Cell membrane / Location in cell: Cell membrane Mesorhizobium loti (bacteria) / Strain: MloK1 / synonym: Soil bacteria / Tissue: Prokaryote / Organelle: Cell membrane / Location in cell: Cell membrane |
| Molecular weight | Experimental: 37.74 KDa / Theoretical: 38.42 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: Cyclic nucleotide
| Macromolecule | Name: Cyclic nucleotide / type: ligand / ID: 2 / Name.synonym: cAMP Details: The ligand for cyclic nucleotide-modulated channel, MloK1 Recombinant expression: No |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Experimental: 369.2 Da / Theoretical: 369.2 Da |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 100 mM KCl, 20 mM Tris-Cl, 5 mM DM, 200 uM cAMP |
| Staining | Type: NEGATIVE Details: Grids with absorbed proteins deeply stained with 2% uranyl formate |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100F |
|---|---|
| Temperature | Average: 298.15 K |
| Date | Oct 27, 2005 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Digitization - Sampling interval: 10 µm / Number real images: 34 / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 60000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side-entry room temperature holder / Specimen holder model: HOME BUILD |
- Image processing
Image processing
| CTF correction | Details: CTF correction of each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C4 (4 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 16.3 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER, EMAN / Number images used: 14921 |
| Final angle assignment | Details: Refinement with MRA method built in EMAN package |
| Final two d classification | Number classes: 190 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)