+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
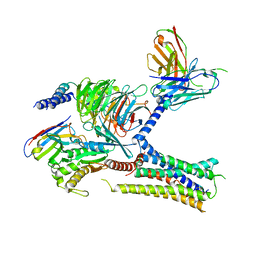
| Title | Structure of the V2 receptor Cter-arrestin2-ScFv30 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G-protein coupled receptor V2 receptor Arrestin 2 Vasopressin / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationangiotensin receptor binding / TGFBR3 regulates TGF-beta signaling / Activation of SMO / negative regulation of interleukin-8 production / G protein-coupled receptor internalization / arrestin family protein binding / Lysosome Vesicle Biogenesis / negative regulation of NF-kappaB transcription factor activity / positive regulation of cardiac muscle hypertrophy / Golgi Associated Vesicle Biogenesis ...angiotensin receptor binding / TGFBR3 regulates TGF-beta signaling / Activation of SMO / negative regulation of interleukin-8 production / G protein-coupled receptor internalization / arrestin family protein binding / Lysosome Vesicle Biogenesis / negative regulation of NF-kappaB transcription factor activity / positive regulation of cardiac muscle hypertrophy / Golgi Associated Vesicle Biogenesis / stress fiber assembly / positive regulation of Rho protein signal transduction / pseudopodium / negative regulation of interleukin-6 production / positive regulation of receptor internalization / enzyme inhibitor activity / negative regulation of Notch signaling pathway / Activated NOTCH1 Transmits Signal to the Nucleus / insulin-like growth factor receptor binding / clathrin-coated pit / negative regulation of protein ubiquitination / GTPase activator activity / cytoplasmic vesicle membrane / Signaling by high-kinase activity BRAF mutants / G protein-coupled receptor binding / MAP2K and MAPK activation / endocytic vesicle membrane / positive regulation of protein phosphorylation / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / Cargo recognition for clathrin-mediated endocytosis / Signaling by BRAF and RAF1 fusions / Clathrin-mediated endocytosis / protein transport / Thrombin signalling through proteinase activated receptors (PARs) / ubiquitin-dependent protein catabolic process / cytoplasmic vesicle / G alpha (s) signalling events / molecular adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / transcription coactivator activity / positive regulation of ERK1 and ERK2 cascade / Ub-specific processing proteases / protein ubiquitination / nuclear body / Golgi membrane / lysosomal membrane / ubiquitin protein ligase binding / regulation of transcription by RNA polymerase II / chromatin / signal transduction / positive regulation of transcription by RNA polymerase II / nucleoplasm / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.23 Å | |||||||||
 Authors Authors | Bous J / Fouillen A / Trapani S / Granier S / Mouillac B / Bron P | |||||||||
| Funding support |  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex. Authors: Bous J / Fouillen A / Orcel H / Trapani S / Cong X / Fontanel S / Saint-Paul J / Lai-Kee-Him J / Urbach S / Sibille N / Sounier R / Granier S / Mouillac B / Bron P | |||||||||
| History |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14223.map.gz emd_14223.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14223-v30.xml emd-14223-v30.xml emd-14223.xml emd-14223.xml | 23.9 KB 23.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14223_fsc.xml emd_14223_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14223.png emd_14223.png | 89.2 KB | ||
| Masks |  emd_14223_msk_1.map emd_14223_msk_1.map | 10.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14223.cif.gz emd-14223.cif.gz | 7.1 KB | ||
| Others |  emd_14223_additional_1.map.gz emd_14223_additional_1.map.gz emd_14223_half_map_1.map.gz emd_14223_half_map_1.map.gz emd_14223_half_map_2.map.gz emd_14223_half_map_2.map.gz | 5 MB 9.7 MB 9.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14223 http://ftp.pdbj.org/pub/emdb/structures/EMD-14223 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14223 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14223 | HTTPS FTP |
-Validation report
| Summary document |  emd_14223_validation.pdf.gz emd_14223_validation.pdf.gz | 824.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14223_full_validation.pdf.gz emd_14223_full_validation.pdf.gz | 823.7 KB | Display | |
| Data in XML |  emd_14223_validation.xml.gz emd_14223_validation.xml.gz | 11.5 KB | Display | |
| Data in CIF |  emd_14223_validation.cif.gz emd_14223_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14223 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14223 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14223 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14223 | HTTPS FTP |
-Related structure data
| Related structure data |  7r0jMC  7r0cC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14223.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14223.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.28 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Ternary complex of the Cterminal portion of the V2 receptor with ...
| Entire | Name: Ternary complex of the Cterminal portion of the V2 receptor with arrestin2 and ScfV30 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of the Cterminal portion of the V2 receptor with ...
| Supramolecule | Name: Ternary complex of the Cterminal portion of the V2 receptor with arrestin2 and ScfV30 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: V2R Cter
| Macromolecule | Name: V2R Cter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.780248 KDa |
| Recombinant expression | Organism: Insect expression vector pBlueBacmsGCA1 (others) |
| Sequence | String: E(SEP)C(TPO)(TPO)A(SEP)(SEP)(SEP)L AKD |
-Macromolecule #2: Arrestin2
| Macromolecule | Name: Arrestin2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.164609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGASWSHPQF EKGGGSGGGS GGSSAWSHPQ FEKLEVLFQG PASGDKGTRV FKKASPNGKL TVYLGKRDFV DHIDLVDPVD GVVLVDPEY LKERRVYVTL TCAFRYGRED LDVLGLTFRK DLFVANVQSF PPAPEDKKPL TRLQERLIKK LGEHAYPFTF E IPPNLPCS ...String: MGASWSHPQF EKGGGSGGGS GGSSAWSHPQ FEKLEVLFQG PASGDKGTRV FKKASPNGKL TVYLGKRDFV DHIDLVDPVD GVVLVDPEY LKERRVYVTL TCAFRYGRED LDVLGLTFRK DLFVANVQSF PPAPEDKKPL TRLQERLIKK LGEHAYPFTF E IPPNLPCS VTLQPGPEDT GKACGVDYEV KAFCAENLEE KIHKRNSVRL VIRKVQYAPE RPGPQPTAET TRQFLMSDKP LH LEASLDK EIYYHGEPIS VNVHVTNNTN KTVKKIKISV RQYADICLFN TAQYKCPVAM EEADDTVAPS STFCKVYTLT PFL ANNREK RGLALDGKLK HEDTNLASST LLREGANREI LGIIVSYKVK VKLVVSRGGL LGDLASSDVA VELPFTLMHP KPKE EPPHR EVPENETPVD TNLIELDTN UniProtKB: Beta-arrestin-1 |
-Macromolecule #3: ScFv30
| Macromolecule | Name: ScFv30 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 30.070027 KDa |
| Recombinant expression | Organism: Insect expression vector pBlueBacmsGCA1 (others) |
| Sequence | String: AMGDIQMTQS PSSLSASVGD RVTITCRASQ SVSSAVAWYQ QKPGKAPKLL IYSASSLYSG VPSRFSGSRS GTDFTLTISS LQPEDFATY YCQQYKYVPV TFGCGTKVEI KGTTAASGSS GGSSSGAEVQ LVESGGGLVQ PGGSLRLSCA ASGFNVYSSS I HWVRQAPG ...String: AMGDIQMTQS PSSLSASVGD RVTITCRASQ SVSSAVAWYQ QKPGKAPKLL IYSASSLYSG VPSRFSGSRS GTDFTLTISS LQPEDFATY YCQQYKYVPV TFGCGTKVEI KGTTAASGSS GGSSSGAEVQ LVESGGGLVQ PGGSLRLSCA ASGFNVYSSS I HWVRQAPG KCLEWVASIS SYYGYTYYAD SVKGRFTISA DTSKNTAYLQ MNSLRAEDTA VYYCARSRQF WYSGLDYWGQ GT LVTVSSA AADDDDKAGW SHPQFEKGGG SGGGSGGGSW SHPQFEK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 14080 / Average exposure time: 1.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller