[English] 日本語
 Yorodumi
Yorodumi- EMDB-14151: BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant fo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant forming a single protofilament (Prosthecobacter dejongeii) | |||||||||
 Map data Map data | BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant forming a single protofilament | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cytyoskeleton / tubulin-like / cytomotive filaments / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule-based process / structural constituent of cytoskeleton / microtubule / hydrolase activity / GTPase activity / GTP binding Similarity search - Function | |||||||||
| Biological species |   Prosthecobacter dejongeii (bacteria) Prosthecobacter dejongeii (bacteria) | |||||||||
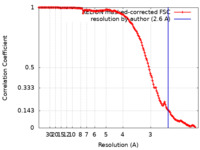
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Wagstaff J / Planelles-Herrero VJ | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics. Authors: James Mark Wagstaff / Vicente José Planelles-Herrero / Grigory Sharov / Aisha Alnami / Frank Kozielski / Emmanuel Derivery / Jan Löwe /  Abstract: Protein filaments are used in myriads of ways to organize other molecules within cells. Some filament-forming proteins couple the hydrolysis of nucleotides to their polymerization cycle, thus ...Protein filaments are used in myriads of ways to organize other molecules within cells. Some filament-forming proteins couple the hydrolysis of nucleotides to their polymerization cycle, thus powering the movement of other molecules. These filaments are termed cytomotive. Only members of the actin and tubulin protein superfamilies are known to form cytomotive filaments. We examined the basis of cytomotivity via structural studies of the polymerization cycles of actin and tubulin homologs from across the tree of life. We analyzed published data and performed structural experiments designed to disentangle functional components of these complex filament systems. Our analysis demonstrates the existence of shared subunit polymerization switches among both cytomotive actins and tubulins, i.e., the conformation of subunits switches upon assembly into filaments. These cytomotive switches can explain filament robustness, by enabling the coupling of kinetic and structural polarities required for cytomotive behaviors and by ensuring that single cytomotive filaments do not fall apart. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14151.map.gz emd_14151.map.gz | 226.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14151-v30.xml emd-14151-v30.xml emd-14151.xml emd-14151.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14151_fsc.xml emd_14151_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14151.png emd_14151.png | 81.3 KB | ||
| Filedesc metadata |  emd-14151.cif.gz emd-14151.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14151 http://ftp.pdbj.org/pub/emdb/structures/EMD-14151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14151 | HTTPS FTP |
-Related structure data
| Related structure data |  7quqMC  7qucC  7qudC  7qupC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14151.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14151.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant forming a single protofilament | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.055 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : BtubAB heterodimer
| Entire | Name: BtubAB heterodimer |
|---|---|
| Components |
|
-Supramolecule #1: BtubAB heterodimer
| Supramolecule | Name: BtubAB heterodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: M-loop mutant BtubA(R284G,K286D,F287G) |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin domain-containing protein
| Macromolecule | Name: Tubulin domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Prosthecobacter dejongeii (bacteria) Prosthecobacter dejongeii (bacteria) |
| Molecular weight | Theoretical: 51.095012 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKVNNTIVVS IGQAGNQIAA SFWKTVCLEH GIDPLTGQTA PGVAPRGNWS SFFSKLGESS SGSYVPRAIM VDLEPSVIDN VKATSGSLF NPANLISRTE GAGGNFAVGY LGAGREVLPE VMSRLDYEID KCDNVGGIIV LHAIGGGTGS GFGALLIESL K EKYGEIPV ...String: MKVNNTIVVS IGQAGNQIAA SFWKTVCLEH GIDPLTGQTA PGVAPRGNWS SFFSKLGESS SGSYVPRAIM VDLEPSVIDN VKATSGSLF NPANLISRTE GAGGNFAVGY LGAGREVLPE VMSRLDYEID KCDNVGGIIV LHAIGGGTGS GFGALLIESL K EKYGEIPV LSCAVLPSPQ VSSVVTEPYN TVFALNTLRR SADACLIFDN EALFDLAHRK WNIESPTVDD LNLLITEALA GI TASMRFS GFLTVEISLR ELLTNLVPQP SLHFLMCAFA PLTPPDGSDG EELGIEEMIK SLFDNGSVFA ACSPMEGRFL STA VLYRGI MEDKPLADAA LAAMREKLPL TYWIPTAFKI GYVEQPGISH RKSMVLLANN TEIARVLDRI CHNFDKLWQR KAFA NWYLN EGMSEEQINV LRASAQELVQ SYQVAEESGA KAKVQDSAGD TGMRAAAAGV SDDARGSMSL RDLVDRRR UniProtKB: Tubulin/FtsZ GTPase domain-containing protein |
-Macromolecule #2: Tubulin beta
| Macromolecule | Name: Tubulin beta / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Prosthecobacter dejongeii (bacteria) Prosthecobacter dejongeii (bacteria) |
| Molecular weight | Theoretical: 46.49757 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MREILSIHVG QCGNQIADSF WRLALREHGL TEAGTLKEGS NAAANSNMEV FFHKVRDGKY VPRAVLVDLE PGVIARIEGG DMSQLFDES SIVRKIPGAA NNWARGYNVE GEKVIDQIMN VIDSAVEKTK GLQGFLMTHS IGGGSGSGLG SLILERLRQA Y PKKRIFTF ...String: MREILSIHVG QCGNQIADSF WRLALREHGL TEAGTLKEGS NAAANSNMEV FFHKVRDGKY VPRAVLVDLE PGVIARIEGG DMSQLFDES SIVRKIPGAA NNWARGYNVE GEKVIDQIMN VIDSAVEKTK GLQGFLMTHS IGGGSGSGLG SLILERLRQA Y PKKRIFTF SVVPSPLISD SAVEPYNAIL TLQRILDNAD GAVLLDNEAL FRIAKAKLNR SPNYMDLNNI IALIVSSVTA SL RFPGKLN TDLSEFVTNL VPFPGNHFLT ASFAPMRGAG QEGQVRTNFP DLARETFAQD NFTAAIDWQQ GVYLAASALF RGD VKAKDV DENMATIRKS LNYASYMPAS GGLKLGYAET APEGFASSGL ALVNHTGIAA VFERLIAQFD IMFDNHAYTH WYEN AGVSR DMMAKARNQI ATLAQSYRDA S UniProtKB: Tubulin beta |
-Macromolecule #3: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER / type: ligand / ID: 3 / Number of copies: 6 / Formula: G2P |
|---|---|
| Molecular weight | Theoretical: 521.208 Da |
| Chemical component information |  ChemComp-G2P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.7 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 6993 / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)