[English] 日本語
 Yorodumi
Yorodumi- EMDB-13935: human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class C -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class C | |||||||||
 Map data Map data | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class C | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Gap junction / Ion Channel / carbon dioxide sensitive / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTransport of connexons to the plasma membrane / gap junction-mediated intercellular transport / gap junction channel activity involved in cell communication by electrical coupling / Oligomerization of connexins into connexons / Transport of connexins along the secretory pathway / gap junction assembly / connexin complex / gap junction / gap junction channel activity / Gap junction assembly ...Transport of connexons to the plasma membrane / gap junction-mediated intercellular transport / gap junction channel activity involved in cell communication by electrical coupling / Oligomerization of connexins into connexons / Transport of connexins along the secretory pathway / gap junction assembly / connexin complex / gap junction / gap junction channel activity / Gap junction assembly / endoplasmic reticulum-Golgi intermediate compartment / sensory perception of sound / transmembrane transport / cell-cell signaling / calcium ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
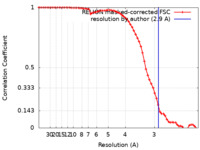
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Brotherton DH / Cameron AD / Savva CG / Ragan TJ | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Conformational changes and CO-induced channel gating in connexin26. Authors: Deborah H Brotherton / Christos G Savva / Timothy J Ragan / Nicholas Dale / Alexander D Cameron /  Abstract: Connexins form large-pore channels that function either as dodecameric gap junctions or hexameric hemichannels to allow the regulated movement of small molecules and ions across cell membranes. ...Connexins form large-pore channels that function either as dodecameric gap junctions or hexameric hemichannels to allow the regulated movement of small molecules and ions across cell membranes. Opening or closing of the channels is controlled by a variety of stimuli, and dysregulation leads to multiple diseases. An increase in the partial pressure of carbon dioxide (PCO) has been shown to cause connexin26 (Cx26) gap junctions to close. Here, we use cryoelectron microscopy (cryo-EM) to determine the structure of human Cx26 gap junctions under increasing levels of PCO. We show a correlation between the level of PCO and the size of the aperture of the pore, governed by the N-terminal helices that line the pore. This indicates that CO alone is sufficient to cause conformational changes in the protein. Analysis of the conformational states shows that movements at the N terminus are linked to both subunit rotation and flexing of the transmembrane helices. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13935.map.gz emd_13935.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13935-v30.xml emd-13935-v30.xml emd-13935.xml emd-13935.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13935_fsc.xml emd_13935_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_13935.png emd_13935.png | 79.2 KB | ||
| Filedesc metadata |  emd-13935.cif.gz emd-13935.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13935 http://ftp.pdbj.org/pub/emdb/structures/EMD-13935 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13935 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13935 | HTTPS FTP |
-Validation report
| Summary document |  emd_13935_validation.pdf.gz emd_13935_validation.pdf.gz | 367.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13935_full_validation.pdf.gz emd_13935_full_validation.pdf.gz | 366.9 KB | Display | |
| Data in XML |  emd_13935_validation.xml.gz emd_13935_validation.xml.gz | 9 KB | Display | |
| Data in CIF |  emd_13935_validation.cif.gz emd_13935_validation.cif.gz | 11.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13935 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13935 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13935 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13935 | HTTPS FTP |
-Related structure data
| Related structure data |  7qeoMC  7qeqC  7qerC  7qesC  7qetC  7qeuC  7qevC  7qewC  7qeyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13935.map.gz / Format: CCP4 / Size: 23.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13935.map.gz / Format: CCP4 / Size: 23.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class C | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
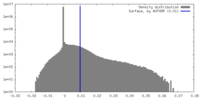
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : two subunits masked from dodecameric assembly of human connexin 2...
| Entire | Name: two subunits masked from dodecameric assembly of human connexin 26: class C |
|---|---|
| Components |
|
-Supramolecule #1: two subunits masked from dodecameric assembly of human connexin 2...
| Supramolecule | Name: two subunits masked from dodecameric assembly of human connexin 26: class C type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gap junction beta-2 protein
| Macromolecule | Name: Gap junction beta-2 protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.713674 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDWGTLQTIL GGVNKHSTSI GKIWLTVLFI FRIMILVVAA KEVWGDEQAD FVCNTLQPGC KNVCYDHYFP ISHIRLWALQ LIFVSTPAL LVAMHVAYRR HEKKRKFIKG EIKSEFKDIE EIKTQKVRIE GSLWWTYTSS IFFRVIFEAA FMYVFYVMYD G FSMQRLVK ...String: MDWGTLQTIL GGVNKHSTSI GKIWLTVLFI FRIMILVVAA KEVWGDEQAD FVCNTLQPGC KNVCYDHYFP ISHIRLWALQ LIFVSTPAL LVAMHVAYRR HEKKRKFIKG EIKSEFKDIE EIKTQKVRIE GSLWWTYTSS IFFRVIFEAA FMYVFYVMYD G FSMQRLVK CNAWPCPNTV DCFVSRPTEK TVFTVFMIAV SGICILLNVT ELCYLLIRYC SGKSKKPVLV PR UniProtKB: Gap junction beta-2 protein |
-Macromolecule #2: DODECYL-BETA-D-MALTOSIDE
| Macromolecule | Name: DODECYL-BETA-D-MALTOSIDE / type: ligand / ID: 2 / Number of copies: 4 / Formula: LMT |
|---|---|
| Molecular weight | Theoretical: 510.615 Da |
| Chemical component information |  ChemComp-LMT: |
-Macromolecule #3: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: The buffer, except DDM and DTT, was prepared fresh from 10x stock on day of use. The basal buffer was filtered and de-gassed, and DDM and DTT added. The buffer was pH corrected at point of ...Details: The buffer, except DDM and DTT, was prepared fresh from 10x stock on day of use. The basal buffer was filtered and de-gassed, and DDM and DTT added. The buffer was pH corrected at point of use to 7.4 using 10% CO2 in 90% N2. | ||||||||||||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: LEICA EM GP Details: 3 microlitres protein applied to grid, blot time 6 seconds, in 10% CO2/90%N2 atmosphere. | ||||||||||||||||||||||||||||||
| Details | This sample monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 4003 / Average exposure time: 5.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)