[English] 日本語
 Yorodumi
Yorodumi- EMDB-13878: Cryo-electron tomogram from a cryo-FIB lift-out lamella of Drosop... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13878 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomogram from a cryo-FIB lift-out lamella of Drosophila melanogaster egg chambers | ||||||||||||
 Map data Map data | Tomogram from cryo-lift-out of D. melanogaster egg chambers | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | cyro-FIB / cryo-electron tomography / D. melanogaster / Golgi apparatus / RIBOSOME | ||||||||||||
| Biological species |  | ||||||||||||
| Method | electron tomography / cryo EM | ||||||||||||
 Authors Authors | Klumpe S / Fung HKH / Goetz SK / Plitzko JM / Mahamid J | ||||||||||||
| Funding support | European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: A modular platform for automated cryo-FIB workflows. Authors: Sven Klumpe / Herman Kh Fung / Sara K Goetz / Ievgeniia Zagoriy / Bernhard Hampoelz / Xiaojie Zhang / Philipp S Erdmann / Janina Baumbach / Christoph W Müller / Martin Beck / Jürgen M ...Authors: Sven Klumpe / Herman Kh Fung / Sara K Goetz / Ievgeniia Zagoriy / Bernhard Hampoelz / Xiaojie Zhang / Philipp S Erdmann / Janina Baumbach / Christoph W Müller / Martin Beck / Jürgen M Plitzko / Julia Mahamid /  Abstract: Lamella micromachining by focused ion beam milling at cryogenic temperature (cryo-FIB) has matured into a preparation method widely used for cellular cryo-electron tomography. Due to the limited ...Lamella micromachining by focused ion beam milling at cryogenic temperature (cryo-FIB) has matured into a preparation method widely used for cellular cryo-electron tomography. Due to the limited ablation rates of low Ga ion beam currents required to maintain the structural integrity of vitreous specimens, common preparation protocols are time-consuming and labor intensive. The improved stability of new-generation cryo-FIB instruments now enables automated operations. Here, we present an open-source software tool, SerialFIB, for creating automated and customizable cryo-FIB preparation protocols. The software encompasses a graphical user interface for easy execution of routine lamellae preparations, a scripting module compatible with available Python packages, and interfaces with three-dimensional correlative light and electron microscopy (CLEM) tools. SerialFIB enables the streamlining of advanced cryo-FIB protocols such as multi-modal imaging, CLEM-guided lamella preparation and in situ lamella lift-out procedures. Our software therefore provides a foundation for further development of advanced cryogenic imaging and sample preparation protocols. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13878.map.gz emd_13878.map.gz | 711.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13878-v30.xml emd-13878-v30.xml emd-13878.xml emd-13878.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13878.png emd_13878.png | 259 KB | ||
| Filedesc metadata |  emd-13878.cif.gz emd-13878.cif.gz | 3.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13878 http://ftp.pdbj.org/pub/emdb/structures/EMD-13878 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13878 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13878 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13878.map.gz / Format: CCP4 / Size: 768.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13878.map.gz / Format: CCP4 / Size: 768.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomogram from cryo-lift-out of D. melanogaster egg chambers | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 14.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
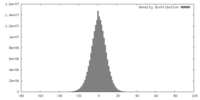
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : D. melanogaster
| Entire | Name: D. melanogaster |
|---|---|
| Components |
|
-Supramolecule #1: D. melanogaster
| Supramolecule | Name: D. melanogaster / type: tissue / ID: 1 / Parent: 0 Details: Tomogram from cryo-FIB lift-out lamella performed on a D. melanogaster egg chamber |
|---|---|
| Source (natural) | Organism:  Strain: w[*]; P{w[+mC]=sqh-mCherry.M}3 (Flybase ID: FBst0059024) Tissue: Egg Chamber |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: NITROGEN / Details: Leica EM Ice. |
| High pressure freezing | Instrument: OTHER Details: 20% ficol in Schneider's medium. The value given for _em_high_pressure_freezing.instrument is EM Ice. This is not in a list of allowed values {'LEICA EM HPM100', 'LEICA EM PACT', 'BAL-TEC ...Details: 20% ficol in Schneider's medium. The value given for _em_high_pressure_freezing.instrument is EM Ice. This is not in a list of allowed values {'LEICA EM HPM100', 'LEICA EM PACT', 'BAL-TEC HPM 010', 'LEICA EM PACT2', 'EMS-002 RAPID IMMERSION FREEZER', 'OTHER'} so OTHER is written into the XML file. |
| Cryo protectant | Ficol |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 / Focused ion beam - Current: 1 / Focused ion beam - Duration: 60 / Focused ion beam - Temperature: 93 K / Focused ion beam - Initial thickness: 10000 / Focused ion beam - Final thickness: 200 Focused ion beam - Details: Cryo-FIB Lift-Out method Various milling steps. The value given for _em_focused_ion_beam.instrument is TFS Aquilos. This is not in a list of allowed values {'DB235', ...Focused ion beam - Details: Cryo-FIB Lift-Out method Various milling steps. The value given for _em_focused_ion_beam.instrument is TFS Aquilos. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD / Number images used: 59 IMOD / Number images used: 59 |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)