[English] 日本語
 Yorodumi
Yorodumi- EMDB-13860: Structure of Hedgehog acyltransferase (HHAT) in complex with mega... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13860 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to IMP-1575 | |||||||||||||||
 Map data Map data | Locally filtered, globally sharpened, NU-refinement focused on the HHAT-Nanobody part | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | HHAT / inhibitor / palmitoyl-CoA / co enzyme A / Hedgehog acyl transferase / Sonic Hedgehog / SHH / MBOAT / morphogen / palmitoylation / signalling / endoplasmic reticulum / membrane protein / heme / small molecule binding / drug target | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationN-terminal peptidyl-L-cysteine N-palmitoylation / O-acyltransferase activity / HHAT G278V doesn't palmitoylate Hh-Np / palmitoyltransferase activity / smoothened signaling pathway / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / Hedgehog ligand biogenesis / Golgi membrane / endoplasmic reticulum membrane / GTP binding ...N-terminal peptidyl-L-cysteine N-palmitoylation / O-acyltransferase activity / HHAT G278V doesn't palmitoylate Hh-Np / palmitoyltransferase activity / smoothened signaling pathway / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / Hedgehog ligand biogenesis / Golgi membrane / endoplasmic reticulum membrane / GTP binding / endoplasmic reticulum / Golgi apparatus Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||||||||
 Authors Authors | Coupland C / Carrique L | |||||||||||||||
| Funding support | European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Structure, mechanism, and inhibition of Hedgehog acyltransferase. Authors: Claire E Coupland / Sebastian A Andrei / T Bertie Ansell / Loic Carrique / Pramod Kumar / Lea Sefer / Rebekka A Schwab / Eamon F X Byrne / Els Pardon / Jan Steyaert / Anthony I Magee / ...Authors: Claire E Coupland / Sebastian A Andrei / T Bertie Ansell / Loic Carrique / Pramod Kumar / Lea Sefer / Rebekka A Schwab / Eamon F X Byrne / Els Pardon / Jan Steyaert / Anthony I Magee / Thomas Lanyon-Hogg / Mark S P Sansom / Edward W Tate / Christian Siebold /   Abstract: The Sonic Hedgehog (SHH) morphogen pathway is fundamental for embryonic development and stem cell maintenance and is implicated in various cancers. A key step in signaling is transfer of a palmitate ...The Sonic Hedgehog (SHH) morphogen pathway is fundamental for embryonic development and stem cell maintenance and is implicated in various cancers. A key step in signaling is transfer of a palmitate group to the SHH N terminus, catalyzed by the multi-pass transmembrane enzyme Hedgehog acyltransferase (HHAT). We present the high-resolution cryo-EM structure of HHAT bound to substrate analog palmityl-coenzyme A and a SHH-mimetic megabody, revealing a heme group bound to HHAT that is essential for HHAT function. A structure of HHAT bound to potent small-molecule inhibitor IMP-1575 revealed conformational changes in the active site that occlude substrate binding. Our multidisciplinary analysis provides a detailed view of the mechanism by which HHAT adapts the membrane environment to transfer an acyl chain across the endoplasmic reticulum membrane. This structure of a membrane-bound O-acyltransferase (MBOAT) superfamily member provides a blueprint for other protein-substrate MBOATs and a template for future drug discovery. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13860.map.gz emd_13860.map.gz | 706.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13860-v30.xml emd-13860-v30.xml emd-13860.xml emd-13860.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13860.png emd_13860.png | 103.5 KB | ||
| Masks |  emd_13860_msk_1.map emd_13860_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13860.cif.gz emd-13860.cif.gz | 7.5 KB | ||
| Others |  emd_13860_half_map_1.map.gz emd_13860_half_map_1.map.gz emd_13860_half_map_2.map.gz emd_13860_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13860 http://ftp.pdbj.org/pub/emdb/structures/EMD-13860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13860 | HTTPS FTP |
-Validation report
| Summary document |  emd_13860_validation.pdf.gz emd_13860_validation.pdf.gz | 613.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13860_full_validation.pdf.gz emd_13860_full_validation.pdf.gz | 612.6 KB | Display | |
| Data in XML |  emd_13860_validation.xml.gz emd_13860_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_13860_validation.cif.gz emd_13860_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13860 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13860 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13860 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13860 | HTTPS FTP |
-Related structure data
| Related structure data |  7q6zMC  7q1uC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13860.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13860.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally filtered, globally sharpened, NU-refinement focused on the HHAT-Nanobody part | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.108 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13860_msk_1.map emd_13860_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13860_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13860_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hedgehog acyltransferase (HHAT) bound to non hydrolysable palmito...
| Entire | Name: Hedgehog acyltransferase (HHAT) bound to non hydrolysable palmitoyl-CoA in complex with megabody 177 |
|---|---|
| Components |
|
-Supramolecule #1: Hedgehog acyltransferase (HHAT) bound to non hydrolysable palmito...
| Supramolecule | Name: Hedgehog acyltransferase (HHAT) bound to non hydrolysable palmitoyl-CoA in complex with megabody 177 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Megabody 177
| Supramolecule | Name: Megabody 177 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Hedgehog acyltransferase (HHAT) bound to non hydrolysable palmito...
| Supramolecule | Name: Hedgehog acyltransferase (HHAT) bound to non hydrolysable palmitoyl-CoA type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Protein-cysteine N-palmitoyltransferase HHAT
| Macromolecule | Name: Protein-cysteine N-palmitoyltransferase HHAT / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.327754 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLPRWELALY LLASLGFHFY SFYEVYKVSR EHEEELDQEF ELETDTLFGG LKKDATDFEW SFWMEWGKQW LVWLLLGHMV VSQMATLLA RKHRPWILML YGMWACWCVL GTPGVAMVLL HTTISFCVAQ FRSQLLTWLC SLLLLSTLRL QGVEEVKRRW Y KTENEYYL ...String: MLPRWELALY LLASLGFHFY SFYEVYKVSR EHEEELDQEF ELETDTLFGG LKKDATDFEW SFWMEWGKQW LVWLLLGHMV VSQMATLLA RKHRPWILML YGMWACWCVL GTPGVAMVLL HTTISFCVAQ FRSQLLTWLC SLLLLSTLRL QGVEEVKRRW Y KTENEYYL LQFTLTVRCL YYTNFSLELC WQQLPAASTS YSFPWMLAYV FYYPVLHNGP ILSFSEFIKQ MQQQEHDSLK AS LCVLALG LGRLLCWWWL AELMAHLMYM HAIYSSIPLL ETVSCWTLGG LALAQVLFFY VKYLVLFGVP ALLMRLDGLT PPA LPRCVS TMFSFTGMWR YFDVGLHNFL IRYVYIPVGG SQHGLLGTLF STAMTFAFVS YWHGGYDYLW CWAALNWLGV TVEN GVRRL VETPCIQDSL ARYFSPQARR RFHAALASCS TSMLILSNLV FLGGNEVGKT YWNRIFIQGW PWVTLSVLGF LYCYS HVGI AWAQTYATDG TETSQVAPA UniProtKB: Protein-cysteine N-palmitoyltransferase HHAT |
-Macromolecule #2: Megabody 177
| Macromolecule | Name: Megabody 177 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 102.601586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAQVQLVESG GGLVKEETQS GLNNYARVVE KGQYDSLEIP AQVAASWESG RDDAAVFGFI DKEQLDKYV ANGGKRSDWT VKFAENRSQD GTLLGYSLLQ ESVDQASYMY SDNHYLAEMA TILGKPEEAK RYRQLAQQLA D YINTCMFD ...String: MKYLLPTAAA GLLLLAAQPA MAQVQLVESG GGLVKEETQS GLNNYARVVE KGQYDSLEIP AQVAASWESG RDDAAVFGFI DKEQLDKYV ANGGKRSDWT VKFAENRSQD GTLLGYSLLQ ESVDQASYMY SDNHYLAEMA TILGKPEEAK RYRQLAQQLA D YINTCMFD PTTQFYYDVR IEDKPLANGC AGKPIVERGK GPEGWSPLFN GAATQANADA VVKVMLDPKE FNTFVPLGTA AL TNPAFGA DIYWRGRVWV DQFWFGLKGM ERYGYRDDAL KLADTFFRHA KGLTADGPIQ ENYNPLTGAQ QGAPNFSWSA AHL YMLYND FFRKQASGGG SGGGGSGGGG SGNADNYKNV INRTGAPQYM KDYDYDDHQR FNPFFDLGAW HGHLLPDGPN TMGG FPGVA LLTEEYINFM ASNFDRLTVW QDGKKVDFTL EAYSIPGALV QKLTAKDVQV EMTLRFATPR TSLLETKITS NKPLD LVWD GELLEKLEAK EGKPLSDKTI AGEYPDYQRK ISATRDGLKV TFGKVRATWD LLTSGESEYQ VHKSLPVQTE INGNRF TSK AHINGSTTLY TTYSHLLTAQ EVSKEQMQIR DILARPAFYL TASQQRWEEY LKKGLTNPDA TPEQTRVAVK AIETLNG NW RSPGGAVKFN TVTPSVTGRW FSGNQTWPWD TWKQAFAMAH FNPDIAKENI RAVFSWQIQP GDSVRPQDVG FVPDLIAW N LSPERGGDGG NWNERNTKPS LAAWSVMEVY NVTQDKTWVA EMYPKLVAYH DWWLRNRDHN GNGVPEYGAT RDKAHNTES GEMLFTVKKS LRLSCTASGA IFSTYDVSWY RQAPEKPREL VAIITRGGNT HYADTVKGRF TISRDNAKKT VNLQMNSLKP EDTAVYYCH AGVQGAMLGP RNYWGQGTQV TVSSHHHHHH |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #4: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 4 / Number of copies: 1 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #5: 2-(2-methylpropylamino)-1-[(4R)-4-(6-methylpyridin-2-yl)-6,7-dihy...
| Macromolecule | Name: 2-(2-methylpropylamino)-1-[(4R)-4-(6-methylpyridin-2-yl)-6,7-dihydro-4H-thieno[3,2-c]pyridin-5-yl]ethanone type: ligand / ID: 5 / Number of copies: 1 / Formula: 9V3 |
|---|---|
| Molecular weight | Theoretical: 343.486 Da |
| Chemical component information | 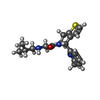 ChemComp-9V3: |
-Macromolecule #6: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 6 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 10683 / Average electron dose: 53.48 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 56 |
|---|---|
| Output model |  PDB-7q6z: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)