+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13654 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DNA polymerase from M. tuberculosis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inhibitor / DNA Polymerase / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology information3'-5' exonuclease activity / DNA-directed DNA polymerase / nucleic acid binding / DNA-directed DNA polymerase activity / DNA replication / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
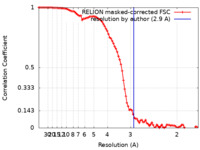
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Borsellini A / Lamers MH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: ACS Infect Dis / Year: 2022 Journal: ACS Infect Dis / Year: 2022Title: DNA-Dependent Binding of Nargenicin to DnaE1 Inhibits Replication in . Authors: Melissa D Chengalroyen / Mandy K Mason / Alessandro Borsellini / Raffaella Tassoni / Garth L Abrahams / Sasha Lynch / Yong-Mo Ahn / Jon Ambler / Katherine Young / Brendan M Crowley / David B ...Authors: Melissa D Chengalroyen / Mandy K Mason / Alessandro Borsellini / Raffaella Tassoni / Garth L Abrahams / Sasha Lynch / Yong-Mo Ahn / Jon Ambler / Katherine Young / Brendan M Crowley / David B Olsen / Digby F Warner / Clifton E Barry Iii / Helena I M Boshoff / Meindert H Lamers / Valerie Mizrahi /    Abstract: Natural products provide a rich source of potential antimicrobials for treating infectious diseases for which drug resistance has emerged. Foremost among these diseases is tuberculosis. Assessment of ...Natural products provide a rich source of potential antimicrobials for treating infectious diseases for which drug resistance has emerged. Foremost among these diseases is tuberculosis. Assessment of the antimycobacterial activity of nargenicin, a natural product that targets the replicative DNA polymerase of , revealed that it is a bactericidal genotoxin that induces a DNA damage response in () and inhibits growth by blocking the replicative DNA polymerase, DnaE1. Cryo-electron microscopy revealed that binding of nargenicin to DnaE1 requires the DNA substrate such that nargenicin is wedged between the terminal base pair and the polymerase and occupies the position of both the incoming nucleotide and templating base. Comparative analysis across three bacterial species suggests that the activity of nargenicin is partly attributable to the DNA binding affinity of the replicative polymerase. This work has laid the foundation for target-led drug discovery efforts focused on DnaE1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13654.map.gz emd_13654.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13654-v30.xml emd-13654-v30.xml emd-13654.xml emd-13654.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13654_fsc.xml emd_13654_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13654.png emd_13654.png | 184.3 KB | ||
| Filedesc metadata |  emd-13654.cif.gz emd-13654.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13654 http://ftp.pdbj.org/pub/emdb/structures/EMD-13654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13654 | HTTPS FTP |
-Related structure data
| Related structure data |  7pu7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13654.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13654.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.866 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
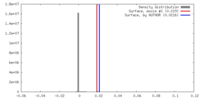
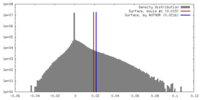
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA polymerase from M. tuberculosis
| Entire | Name: DNA polymerase from M. tuberculosis |
|---|---|
| Components |
|
-Supramolecule #1: DNA polymerase from M. tuberculosis
| Supramolecule | Name: DNA polymerase from M. tuberculosis / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 129.3 kDa/nm |
-Macromolecule #1: DNA polymerase III subunit alpha
| Macromolecule | Name: DNA polymerase III subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 129.479734 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MSGSSAGSSF VHLHNHTEYS MLDGAAKITP MLAEVERLGM PAVGMTDHGN MFGASEFYNS ATKAGIKPII GVEAYIAPGS RFDTRRILW GDPSQKADDV SGSGSYTHLT MMAENATGLR NLFKLSSHAS FEGQLSKWSR MDAELIAEHA EGIIITTGCP S GEVQTRLR ...String: MSGSSAGSSF VHLHNHTEYS MLDGAAKITP MLAEVERLGM PAVGMTDHGN MFGASEFYNS ATKAGIKPII GVEAYIAPGS RFDTRRILW GDPSQKADDV SGSGSYTHLT MMAENATGLR NLFKLSSHAS FEGQLSKWSR MDAELIAEHA EGIIITTGCP S GEVQTRLR LGQDREALEA AAKWREIVGP DNYFLELMDH GLTIERRVRD GLLEIGRALN IPPLATNDCH YVTRDAAHNH EA LLCVQTG KTLSDPNRFK FDGDGYYLKS AAEMRQIWDD EVPGACDSTL LIAERVQSYA DVWTPRDRMP VFPVPDGHDQ ASW LRHEVD AGLRRRFPAG PPDGYRERAA YEIDVICSKG FPSYFLIVAD LISYARSAGI RVGPGRGSAA GSLVAYALGI TDID PIPHG LLFERFLNPE RTSMPDIDID FDDRRRGEMV RYAADKWGHD RVAQVITFGT IKTKAALKDS ARIHYGQPGF AIADR ITKA LPPAIMAKDI PLSGITDPSH ERYKEAAEVR GLIETDPDVR TIYQTARGLE GLIRNAGVHA CAVIMSSEPL TEAIPL WKR PQDGAIITGW DYPACEAIGL LKMDFLGLRN LTIIGDAIDN VRANRGIDLD LESVPLDDKA TYELLGRGDT LGVFQLD GG PMRDLLRRMQ PTGFEDVVAV IALYRPGPMG MNAHNDYADR KNNRQAIKPI HPELEEPLRE ILAETYGLIV YQEQIMRI A QKVASYSLAR ADILRKAMGK KKREVLEKEF EGFSDGMQAN GFSPAAIKAL WDTILPFADY AFNKSHAAGY GMVSYWTAY LKANYPAEYM AGLLTSVGDD KDKAAVYLAD CRKLGITVLP PDVNESGLNF ASVGQDIRYG LGAVRNVGAN VVGSLLQTRN DKGKFTDFS DYLNKIDISA CNKKVTESLI KAGAFDSLGH ARKGLFLVHS DAVDSVLGTK KAEALGQFDL FGSNDDGTGT A DPVFTIKV PDDEWEDKHK LALEREMLGL YVSGHPLNGV AHLLAAQVDT AIPAILDGDV PNDAQVRVGG ILASVNRRVN KN GMPWASA QLEDLTGGIE VMFFPHTYSS YGADIVDDAV VLVNAKVAVR DDRIALIAND LTVPDFSNAE VERPLAVSLP TRQ CTFDKV SALKQVLARH PGTSQVHLRL ISGDRITTLA LDQSLRVTPS PALMGDLKEL LGPGCLGS UniProtKB: DNA polymerase III subunit alpha |
-Macromolecule #2: Template
| Macromolecule | Name: Template / type: other / ID: 2 / Number of copies: 1 Classification: polydeoxyribonucleotide/polyribonucleotide hybrid |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 5.944902 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DG)(DA)(DA)(DG)(DG)(DA) (DC)(DG)(DA)(DA)(DG)(DG)(DA)(DC)(DT) |
-Macromolecule #3: primer
| Macromolecule | Name: primer / type: other / ID: 3 / Number of copies: 1 Classification: polydeoxyribonucleotide/polyribonucleotide hybrid |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.197727 KDa |
| Sequence | String: (DA)(DG)(DT)(DC)(DC)(DT)(DT)(DC)(DG)(DT) (DC)(DC)(DT)(DT) |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: [(1~{S},3~{R},4~{R},5~{R},6~{R},7~{S},8~{R},11~{S},13~{S},16~{S},...
| Macromolecule | Name: [(1~{S},3~{R},4~{R},5~{R},6~{R},7~{S},8~{R},11~{S},13~{S},16~{S},17~{R},18~{E})-13-methoxy-5,17,19-trimethyl-6-oxidanyl-16-[(1~{R})-1-oxidanylethyl]-14-oxidanylidene-2,15-dioxatetracyclo[9.8.0. ...Name: [(1~{S},3~{R},4~{R},5~{R},6~{R},7~{S},8~{R},11~{S},13~{S},16~{S},17~{R},18~{E})-13-methoxy-5,17,19-trimethyl-6-oxidanyl-16-[(1~{R})-1-oxidanylethyl]-14-oxidanylidene-2,15-dioxatetracyclo[9.8.0.0^{1,7}.0^{3,8}]nonadeca-9,18-dien-4-yl] 1~{H}-pyrrole-2-carboxylate type: ligand / ID: 5 / Number of copies: 1 / Formula: 82W |
|---|---|
| Molecular weight | Theoretical: 515.595 Da |
| Chemical component information |  ChemComp-82W: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 3 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.0002 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 76 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7pu7: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)