+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13639 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In-vitro structure of inverted S-layer tube | |||||||||
 Map data Map data | Postprocessed map without B-factor sharpening | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Surface glycoprotein signal peptide / Major cell surface glycoprotein / PGF-CTERM archaeal protein-sorting signal / PGF-CTERM motif / S-layer / cell wall organization / extracellular region / plasma membrane / Cell surface glycoprotein Function and homology information Function and homology information | |||||||||
| Biological species |  Haloferax volcanii DS2 (archaea) Haloferax volcanii DS2 (archaea) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 15.87 Å | |||||||||
 Authors Authors | von Kuegelgen A / Bharat TAM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Complete atomic structure of a native archaeal cell surface. Authors: Andriko von Kügelgen / Vikram Alva / Tanmay A M Bharat /   Abstract: Many prokaryotic cells are covered by an ordered, proteinaceous, sheet-like structure called a surface layer (S-layer). S-layer proteins (SLPs) are usually the highest copy number macromolecules in ...Many prokaryotic cells are covered by an ordered, proteinaceous, sheet-like structure called a surface layer (S-layer). S-layer proteins (SLPs) are usually the highest copy number macromolecules in prokaryotes, playing critical roles in cellular physiology such as blocking predators, scaffolding membranes, and facilitating environmental interactions. Using electron cryomicroscopy of two-dimensional sheets, we report the atomic structure of the S-layer from the archaeal model organism Haloferax volcanii. This S-layer consists of a hexagonal array of tightly interacting immunoglobulin-like domains, which are also found in SLPs across several classes of archaea. Cellular tomography reveal that the S-layer is nearly continuous on the cell surface, completed by pentameric defects in the hexagonal lattice. We further report the atomic structure of the SLP pentamer, which shows markedly different relative arrangements of SLP domains needed to complete the S-layer. Our structural data provide a framework for understanding cell surfaces of archaea at the atomic level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13639.map.gz emd_13639.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13639-v30.xml emd-13639-v30.xml emd-13639.xml emd-13639.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13639.png emd_13639.png | 88.9 KB | ||
| Masks |  emd_13639_msk_1.map emd_13639_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_13639_additional_1.map.gz emd_13639_additional_1.map.gz | 5.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13639 http://ftp.pdbj.org/pub/emdb/structures/EMD-13639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13639 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13639.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13639.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map without B-factor sharpening | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.238 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
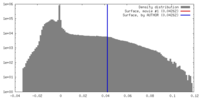
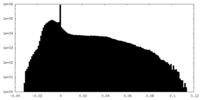
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13639_msk_1.map emd_13639_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
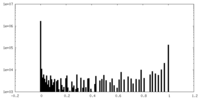
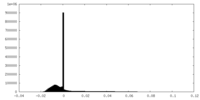
| Density Histograms |
-Additional map: Full map without post-processing and B-factor sharpening
| File | emd_13639_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map without post-processing and B-factor sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : In-vitro structure of inverted S-layer tube
| Entire | Name: In-vitro structure of inverted S-layer tube |
|---|---|
| Components |
|
-Supramolecule #1: In-vitro structure of inverted S-layer tube
| Supramolecule | Name: In-vitro structure of inverted S-layer tube / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: In-vitro structure of inverted S-layer tube |
|---|---|
| Source (natural) | Organism:  Haloferax volcanii DS2 (archaea) / Location in cell: Cell surface Haloferax volcanii DS2 (archaea) / Location in cell: Cell surface |
-Macromolecule #1: S-layer protein
| Macromolecule | Name: S-layer protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloferax volcanii DS2 (archaea) / Strain: H26 Haloferax volcanii DS2 (archaea) / Strain: H26 |
| Sequence | String: ERGNLDADSE SFNKTIQSGD RVFLGE EIS TDAGLGASNP LLTGTAGNSE GVSLDLSSPI PQTTENQPLG TYDVDGSGSA TTPNVTL LA PRITDSEILT SSGGDVTGSA ISSSDAGNLY VNADYNYESA EKVEVTVEDP SGTDITNE V LSGTDTFVDD GSIGSTSSTG ...String: ERGNLDADSE SFNKTIQSGD RVFLGE EIS TDAGLGASNP LLTGTAGNSE GVSLDLSSPI PQTTENQPLG TYDVDGSGSA TTPNVTL LA PRITDSEILT SSGGDVTGSA ISSSDAGNLY VNADYNYESA EKVEVTVEDP SGTDITNE V LSGTDTFVDD GSIGSTSSTG GGVGIDMSDQ DAGEYTIILE GAEDLDFGDA TETMTLTIS SQDEIGIELD SESVTQGTDV QYTVTNGIDG NEHVVAMDLS DLQNDATTEQ AKEVFRNIGD TSEVGIANS SATNTSGSST GPTVETADIA YAVVEIDGAS AVGGIETQYL DDSEVDLEVY D AGVSATAA VGQDATNDIT LTIEEGGTTL SSPTGQYVVG SEVDINGTAT SSDSVAIYVR DD GDWQLLE IGGDNEISVD SDDTFEEEDI ALSGLSGDGS SILSLTGTYR IGVIDASDAD VGG DGSVDD SLTTSEFTSG VSSSNSIRVT DQALTGQFTT INGQVAPVET GTVDINGTAS GANS VLVIF VDERGNVNYQ EVSVDSDGTY DEDDITVGLT QGRVTAHILS VGRDSAIGDG SLPSG PSNG ATLNDLTGYL DTLDQNNNNG EQINELIASE TVDETASDDL IVTETFRLAE SSTSID SIY PDAAEAAGIN PVATGETMVI AGSTNLKPDD NTISIEVTNE DGTSVALEDT DEWNNDG QW MVEIDTTDFE TGTFTVEADD GDNTDTVNVE VVSEREDTTT SSDNATDTTT TTDGPTET T TTAEPTETTE EPTEETTTSS NTPGFGIAVA LVALVGAALL ALRREN |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: Buffer solutions were prepared fresh from sterile filtered concentrated stocksolutions. Solutions were filtered through a 0.22 um filter to avoid microbial contamination and degassed using a ...Details: Buffer solutions were prepared fresh from sterile filtered concentrated stocksolutions. Solutions were filtered through a 0.22 um filter to avoid microbial contamination and degassed using a vacuum fold pump. The pH of the HEPES stock solution was adjusted with sodium hydroxide at 4 deg C. 15 mM Calcium chloride was added 15 minutes before vitrification. | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Atmosphere: AIR / Details: 20 seconds, 15 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: Vitrobot options: Blot time 2.0 seconds, Blot force -15,1, Wait time 0 seconds, Drain time 0.5 seconds. | |||||||||||||||
| Details | Haloferax volcanii vesicles |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Spherical aberration corrector: not used / Chromatic aberration corrector: not used / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Details | SerialEM Dose symmetric scheme |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 2 / Average exposure time: 2.4 sec. / Average electron dose: 3.0 e/Å2 Details: Dose symmetric tilt scheme (Hagen et al, JSB); 2.4 seconds of exposure with 6 saved frames. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 0.4 µm / Calibrated defocus min: 0.1 µm / Calibrated magnification: 64000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Rigid body fit inside coot of D1-D6 domains and real space refinement with restraints of the original model obtained by single particle analysis in PHENIX. |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Best Fit |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)