+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1360 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of TOR and its complex with KOG1. | |||||||||
 Map data Map data | 3D reconstruction of the yeast TOR1(target of rapamycin 1)protein | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | HEAT repeat / 1-phosphatidylinositol-3-kinase activity Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Adami A / Garcia-Alvarez B / Arias-Palomo E / Barford D / Llorca O | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2007 Journal: Mol Cell / Year: 2007Title: Structure of TOR and its complex with KOG1. Authors: Alessandra Adami / Begoña García-Alvarez / Ernesto Arias-Palomo / David Barford / Oscar Llorca /  Abstract: The target of rapamycin (TOR) is a large (281 kDa) conserved Ser/Thr protein kinase that functions as a central controller of cell growth. TOR assembles into two distinct multiprotein complexes: ...The target of rapamycin (TOR) is a large (281 kDa) conserved Ser/Thr protein kinase that functions as a central controller of cell growth. TOR assembles into two distinct multiprotein complexes: TORC1 and TORC2. A defining feature of TORC1 is the interaction of TOR with KOG1 (Raptor in mammals) and its sensitivity to a rapamycin-FKBP12 complex. Here, we have reconstructed in three dimensions the 25 A resolution structures of endogenous budding yeast TOR1 and a TOR-KOG1 complex, using electron microscopy. TOR features distinctive N-terminal HEAT repeats that form a curved tubular-shaped domain that associates with the C-terminal WD40 repeat domain of KOG1. The N terminus of KOG1 is in proximity to the TOR kinase domain, likely functioning to bring substrates into the vicinity of the catalytic region. A model is proposed for the molecular architecture of the TOR-KOG1 complex explaining its sensitivity to rapamycin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1360.map.gz emd_1360.map.gz | 74.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1360-v30.xml emd-1360-v30.xml emd-1360.xml emd-1360.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| Images |  1360.gif 1360.gif | 56.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1360 http://ftp.pdbj.org/pub/emdb/structures/EMD-1360 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1360 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1360 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1360.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1360.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of the yeast TOR1(target of rapamycin 1)protein | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
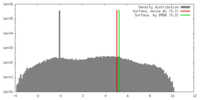
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TOR1
| Entire | Name: TOR1 |
|---|---|
| Components |
|
-Supramolecule #1000: TOR1
| Supramolecule | Name: TOR1 / type: sample / ID: 1000 / Oligomeric state: One monomer of yeast TOR1 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 281 KDa |
-Macromolecule #1: TOR1
| Macromolecule | Name: TOR1 / type: protein_or_peptide / ID: 1 / Name.synonym: target of rapamycin / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 281 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: 1-phosphatidylinositol-3-kinase activity / InterPro: HEAT repeat |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.1 Details: 50mM HEPES-KOH pH7.1, 3mM DTT, 10% glycerol, 0.01% Tween 20 for free TOR1, 150 mM KCl, 1 mM Mg acetate, 2 mM EGTA |
|---|---|
| Staining | Type: NEGATIVE / Details: 1% uranyl acetate |
| Grid | Details: 400 mesh Copper/Palladium grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1230 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: correction with FFT and CCD camera |
| Details | Microscope used - JEOL JEM-1230 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 10.5 µm Details: images scanned with a MINOLTA Dimage Scan Multi Pro scanner at 2400 dpi and averaged to a final 4.2 angstroms per pixel at the specimen Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.9 mm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
- Image processing
Image processing
| CTF correction | Details: CTF for each micrograph was estimated using CTFIND3 and the phases flipped with BSOFT |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 5643 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)