+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lipid Flippase / P4 ATPase / trans-Golgi Network / Phosphatidylserine transport / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationCdc50p-Drs2p complex / actin cortical patch localization / aminophospholipid translocation / phosphatidylcholine flippase activity / Ion transport by P-type ATPases / post-Golgi vesicle-mediated transport / phosphatidylserine flippase activity / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / ATPase-coupled intramembrane lipid transporter activity ...Cdc50p-Drs2p complex / actin cortical patch localization / aminophospholipid translocation / phosphatidylcholine flippase activity / Ion transport by P-type ATPases / post-Golgi vesicle-mediated transport / phosphatidylserine flippase activity / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / ATPase-coupled intramembrane lipid transporter activity / phosphatidylethanolamine flippase activity / endocytic recycling / P-type phospholipid transporter / phosphatidylinositol-4-phosphate binding / retrograde transport, endosome to Golgi / phospholipid translocation / Neutrophil degranulation / intracellular protein transport / trans-Golgi network / endocytosis / late endosome membrane / endosome membrane / magnesium ion binding / endoplasmic reticulum / Golgi apparatus / ATP hydrolysis activity / ATP binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Timcenko M / Wang Y | |||||||||
| Funding support |  Denmark, 1 items Denmark, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Substrate Transport and Specificity in a Phospholipid Flippase Authors: Wang Y / Lyons JA / Timcenko M / Kummerer F / de Groot BL / Nissen P / Gapsys V / Lindorff-Larsen K | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13353.map.gz emd_13353.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13353-v30.xml emd-13353-v30.xml emd-13353.xml emd-13353.xml | 29.8 KB 29.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13353_fsc.xml emd_13353_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_13353.png emd_13353.png | 96 KB | ||
| Filedesc metadata |  emd-13353.cif.gz emd-13353.cif.gz | 8.7 KB | ||
| Others |  emd_13353_additional_1.map.gz emd_13353_additional_1.map.gz emd_13353_half_map_1.map.gz emd_13353_half_map_1.map.gz emd_13353_half_map_2.map.gz emd_13353_half_map_2.map.gz | 32.2 MB 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13353 http://ftp.pdbj.org/pub/emdb/structures/EMD-13353 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13353 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13353 | HTTPS FTP |
-Related structure data
| Related structure data |  7pemMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13353.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13353.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0312 Å | ||||||||||||||||||||||||||||||||||||
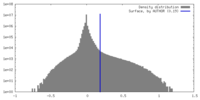
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Map
| File | emd_13353_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
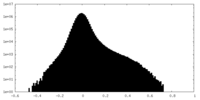
| Density Histograms |
-Half map: Half-map A
| File | emd_13353_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B
| File | emd_13353_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Binary complex of Drs2p-Cdc50p with the regulatory lipid PI4P and...
| Entire | Name: Binary complex of Drs2p-Cdc50p with the regulatory lipid PI4P and transport substrate PS |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of Drs2p-Cdc50p with the regulatory lipid PI4P and...
| Supramolecule | Name: Binary complex of Drs2p-Cdc50p with the regulatory lipid PI4P and transport substrate PS type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Probable phospholipid-transporting ATPase DRS2
| Macromolecule | Name: Probable phospholipid-transporting ATPase DRS2 / type: protein_or_peptide / ID: 1 Details: Drs2p with a Thrombin cleavable C-terminal biotin acceptor domain (BAD) tag, and additional Thrombin cleavage site in the C-terminus. Number of copies: 1 / Enantiomer: LEVO / EC number: P-type phospholipid transporter |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 154.006766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNDDRETPPK RKPGEDDTLF DIDFLDDTTS HSGSRSKVTN SHANANYIPP SHVLPEETID LDADDDNIEN DVHENLFMSN NHDDQTSWN ANRFDSDAYQ PQSLRAVKPP GLFARFGNGL KNAFTFKRKK GPESFEMNHY NAVTNNELDD NYLDSRNKFN I KILFNRYI ...String: MNDDRETPPK RKPGEDDTLF DIDFLDDTTS HSGSRSKVTN SHANANYIPP SHVLPEETID LDADDDNIEN DVHENLFMSN NHDDQTSWN ANRFDSDAYQ PQSLRAVKPP GLFARFGNGL KNAFTFKRKK GPESFEMNHY NAVTNNELDD NYLDSRNKFN I KILFNRYI LRKNVGDAEG NGEPRVIHIN DSLANSSFGY SDNHISTTKY NFATFLPKFL FQEFSKYANL FFLCTSAIQQ VP HVSPTNR YTTIGTLLVV LIVSAMKECI EDIKRANSDK ELNNSTAEIF SEAHDDFVEK RWIDIRVGDI IRVKSEEPIP ADT IILSSS EPEGLCYIET ANLDGETNLK IKQSRVETAK FIDVKTLKNM NGKVVSEQPN SSLYTYEGTM TLNDRQIPLS PDQM ILRGA TLRNTAWIFG LVIFTGHETK LLRNATATPI KRTAVEKIIN RQIIALFTVL IVLILISSIG NVIMSTADAK HLSYL YLEG TNKAGLFFKD FLTFWILFSN LVPISLFVTV ELIKYYQAFM IGSDLDLYYE KTDTPTVVRT SSLVEELGQI EYIFS (PHD)KTG TLTRNIMEFK SCSIAGHCYI DKIPEDKTAT VEDGIEVGYR KFDDLKKKLN DPSDEDSPII NDFLTLLATC HT VIPEFQS DGSIKYQAAS PDEGALVQGG ADLGYKFIIR KPNSVTVLLE ETGEEKEYQL LNICEFNSTR KRMSAIFRFP DGS IKLFCK GADTVILERL DDEANQYVEA TMRHLEDYAS EGLRTLCLAM RDISEGEYEE WNSIYNEAAT TLDNRAEKLD EAAN LIEKN LILIGATAIE DKLQDGVPET IHTLQEAGIK IWVLTGDRQE TAINIGMSCR LLSEDMNLLI INEETRDDTE RNLLE KINA LNEHQLSTHD MNTLALVIDG KSLGFALEPE LEDYLLTVAK LCKAVICCRV SPLQKALVVK MVKRKSSSLL LAIGDG AND VSMIQAAHVG VGISGMEGMQ AARSADIAVG QFKFLKKLLL VHGSWSYQRI SVAILYSFYK NTALYMTQFW YVFANAF SG QSIMESWTMS FYNLFFTVWP PFVIGVFDQF VSSRLLERYP QLYKLGQKGQ FFSVYIFWGW IINGFFHSAI VFIGTILI Y RYGFALNMHG ELADHWSWGV TVYTTSVIIV LGKAALVTNQ WTKFTLIAIP GSLLFWLIFF PIYASIFPHA NISREYYGV VKHTYGSGVF WLTLIVLPIF ALVRDFLWKY YKRMYEPETY HVIQEMQKYN ISDSRPHVQQ FQNAIRKVRQ VQRMKKQRGF AFSQAEEGG QEKIVRMYDT TQKRGKYGEL QDASANPFND NNGLGSNDFE SAEPFIENPF ADGNQNSNRF SSSRDDISFD I UniProtKB: Phospholipid-transporting ATPase DRS2 |
-Macromolecule #2: Cell division control protein 50
| Macromolecule | Name: Cell division control protein 50 / type: protein_or_peptide / ID: 2 / Details: Cdc50p with Thrombin cleavable C-terminal His-tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 45.037312 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVSLFKRGKA PPLTKEGPTS KKPPNTAFRQ QRLKAWQPIL SPQSVLPLLI FVACIFTPIG IGLIVSATKV QDLTIDYSHC DTKASTTAF EDIPKKYIKY HFKSKVENKP QWRLTENENG EQSCELQFEI PNDIKKSIFI YYKITNFYQN HRRYVQSFDT K QILGEPIK ...String: MVSLFKRGKA PPLTKEGPTS KKPPNTAFRQ QRLKAWQPIL SPQSVLPLLI FVACIFTPIG IGLIVSATKV QDLTIDYSHC DTKASTTAF EDIPKKYIKY HFKSKVENKP QWRLTENENG EQSCELQFEI PNDIKKSIFI YYKITNFYQN HRRYVQSFDT K QILGEPIK KDDLDTSCSP IRSREDKIIY PCGLIANSMF NDTFSQVLSG IDDTEDYNLT NKHISWSIDR HRFKTTKYNA SD IVPPPNW MKKYPDGYTD ENLPDIHTWE EFQVWMRTAA FPKFYKLTLK NESASLPKGK YQMNIELNYP ISLFGGTKSF VLT TNGAIG GRNMSLGVLY LIVAGLCALF GIIFLVKLIF QPRAMGDHTY LNFDDEENED YEDVHAENTT LREIL UniProtKB: Phospholipid-transporting ATPase accessory subunit CDC50 |
-Macromolecule #5: (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-...
| Macromolecule | Name: (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate type: ligand / ID: 5 / Number of copies: 1 / Formula: 2Y5 |
|---|---|
| Molecular weight | Theoretical: 967.108 Da |
| Chemical component information |  ChemComp-2Y5: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #8: O-[(R)-[(2S)-2-(hexadecanoyloxy)-3-(octadecanoyloxy)propoxy](hydr...
| Macromolecule | Name: O-[(R)-[(2S)-2-(hexadecanoyloxy)-3-(octadecanoyloxy)propoxy](hydroxy)phosphoryl]-D-serine type: ligand / ID: 8 / Number of copies: 1 / Formula: Q3G |
|---|---|
| Molecular weight | Theoretical: 764.022 Da |
| Chemical component information |  ChemComp-Q3G: |
-Macromolecule #9: water
| Macromolecule | Name: water / type: ligand / ID: 9 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Incubated with 0.1 mg/mL POPS for 1 hour. 3mM ATP was added to the sample just before application to the grid.. | ||||||||||||||||||
| Details | Purified in detergent lauryl maltose neopentyl glycol (LMNG) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 9837 / Average exposure time: 1.5 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Molecular dynamics flexible fitting and energy minimization in Gromacs |
| Refinement | Space: REAL / Overall B value: 69.1 / Target criteria: correlation coefficient |
| Output model |  PDB-7pem: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)