[English] 日本語
 Yorodumi
Yorodumi- EMDB-13348: cryo-EM structure of DEPTOR bound to human mTOR complex 2, focuss... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13348 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
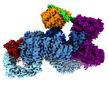
| Title | cryo-EM structure of DEPTOR bound to human mTOR complex 2, focussed on one protomer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of extrinsic apoptotic signaling pathway / RNA polymerase III type 2 promoter sequence-specific DNA binding / RNA polymerase III type 1 promoter sequence-specific DNA binding / positive regulation of cytoplasmic translational initiation / regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / regulation of locomotor rhythm / T-helper 1 cell lineage commitment / positive regulation of pentose-phosphate shunt / positive regulation of wound healing, spreading of epidermal cells / TORC2 signaling ...regulation of extrinsic apoptotic signaling pathway / RNA polymerase III type 2 promoter sequence-specific DNA binding / RNA polymerase III type 1 promoter sequence-specific DNA binding / positive regulation of cytoplasmic translational initiation / regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / regulation of locomotor rhythm / T-helper 1 cell lineage commitment / positive regulation of pentose-phosphate shunt / positive regulation of wound healing, spreading of epidermal cells / TORC2 signaling / TORC2 complex / regulation of membrane permeability / cellular response to leucine starvation / negative regulation of lysosome organization / heart valve morphogenesis / TFIIIC-class transcription factor complex binding / TORC1 complex / voluntary musculoskeletal movement / positive regulation of transcription of nucleolar large rRNA by RNA polymerase I / regulation of cellular response to oxidative stress / negative regulation of TORC2 signaling / calcineurin-NFAT signaling cascade / RNA polymerase III type 3 promoter sequence-specific DNA binding / positive regulation of keratinocyte migration / regulation of osteoclast differentiation / regulation of lysosome organization / MTOR signalling / cellular response to nutrient / cellular response to L-leucine / Amino acids regulate mTORC1 / energy reserve metabolic process / regulation of autophagosome assembly / Energy dependent regulation of mTOR by LKB1-AMPK / phosphatidic acid binding / TORC1 signaling / negative regulation of Ras protein signal transduction / ruffle organization / serine/threonine protein kinase complex / embryo development ending in birth or egg hatching / cellular response to methionine / phosphatidylinositol-3,4-bisphosphate binding / negative regulation of cell size / positive regulation of ubiquitin-dependent protein catabolic process / cellular response to osmotic stress / negative regulation of TOR signaling / negative regulation of protein localization to nucleus / phosphatidylinositol-3,5-bisphosphate binding / anoikis / inositol hexakisphosphate binding / cardiac muscle cell development / negative regulation of calcineurin-NFAT signaling cascade / regulation of myelination / regulation of establishment of cell polarity / positive regulation of transcription by RNA polymerase III / negative regulation of macroautophagy / Macroautophagy / lipid biosynthetic process / positive regulation of myotube differentiation / regulation of cell size / Constitutive Signaling by AKT1 E17K in Cancer / positive regulation of actin filament polymerization / protein kinase inhibitor activity / germ cell development / phosphatidylinositol-3,4,5-trisphosphate binding / TOR signaling / behavioral response to pain / mTORC1-mediated signalling / oligodendrocyte differentiation / positive regulation of oligodendrocyte differentiation / positive regulation of translational initiation / protein serine/threonine kinase inhibitor activity / CD28 dependent PI3K/Akt signaling / HSF1-dependent transactivation / positive regulation of TOR signaling / regulation of macroautophagy / enzyme-substrate adaptor activity / response to amino acid / 'de novo' pyrimidine nucleobase biosynthetic process / positive regulation of epithelial to mesenchymal transition / vascular endothelial cell response to laminar fluid shear stress / positive regulation of lipid biosynthetic process / heart morphogenesis / cellular response to nutrient levels / neuronal action potential / negative regulation of protein kinase activity / regulation of cellular response to heat / positive regulation of lamellipodium assembly / cardiac muscle contraction / phagocytic vesicle / T cell costimulation / positive regulation of stress fiber assembly / negative regulation of TORC1 signaling / positive regulation of endothelial cell proliferation / phosphatidylinositol-4,5-bisphosphate binding / cytoskeleton organization / positive regulation of autophagy / substantia nigra development / endomembrane system / negative regulation of insulin receptor signaling pathway / negative regulation of autophagy Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Waelchli M / Maier T | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Regulation of human mTOR complexes by DEPTOR. Authors: Matthias Wälchli / Karolin Berneiser / Francesca Mangia / Stefan Imseng / Louise-Marie Craigie / Edward Stuttfeld / Michael N Hall / Timm Maier /  Abstract: The vertebrate-specific DEP domain-containing mTOR interacting protein (DEPTOR), an oncoprotein or tumor suppressor, has important roles in metabolism, immunity, and cancer. It is the only protein ...The vertebrate-specific DEP domain-containing mTOR interacting protein (DEPTOR), an oncoprotein or tumor suppressor, has important roles in metabolism, immunity, and cancer. It is the only protein that binds and regulates both complexes of mammalian target of rapamycin (mTOR), a central regulator of cell growth. Biochemical analysis and cryo-EM reconstructions of DEPTOR bound to human mTOR complex 1 (mTORC1) and mTORC2 reveal that both structured regions of DEPTOR, the PDZ domain and the DEP domain tandem (DEPt), are involved in mTOR interaction. The PDZ domain binds tightly with mildly activating effect, but then acts as an anchor for DEPt association that allosterically suppresses mTOR activation. The binding interfaces of the PDZ domain and DEPt also support further regulation by other signaling pathways. A separate, substrate-like mode of interaction for DEPTOR phosphorylation by mTOR complexes rationalizes inhibition of non-stimulated mTOR activity at higher DEPTOR concentrations. The multifaceted interplay between DEPTOR and mTOR provides a basis for understanding the divergent roles of DEPTOR in physiology and opens new routes for targeting the mTOR-DEPTOR interaction in disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13348.map.gz emd_13348.map.gz | 383.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13348-v30.xml emd-13348-v30.xml emd-13348.xml emd-13348.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13348.png emd_13348.png | 135 KB | ||
| Masks |  emd_13348_msk_1.map emd_13348_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13348 http://ftp.pdbj.org/pub/emdb/structures/EMD-13348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13348 | HTTPS FTP |
-Validation report
| Summary document |  emd_13348_validation.pdf.gz emd_13348_validation.pdf.gz | 256.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13348_full_validation.pdf.gz emd_13348_full_validation.pdf.gz | 255.3 KB | Display | |
| Data in XML |  emd_13348_validation.xml.gz emd_13348_validation.xml.gz | 7.8 KB | Display | |
| Data in CIF |  emd_13348_validation.cif.gz emd_13348_validation.cif.gz | 9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13348 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13348 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13348 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13348 | HTTPS FTP |
-Related structure data
| Related structure data |  7pe8MC  7pe7C  7pe9C  7peaC  7pebC  7pecC  7pedC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13348.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13348.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
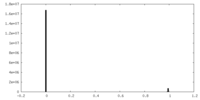
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13348_msk_1.map emd_13348_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
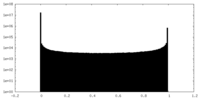
| Density Histograms |
- Sample components
Sample components
-Entire : mTORC2 in complex with its regulator DEPTOR
| Entire | Name: mTORC2 in complex with its regulator DEPTOR |
|---|---|
| Components |
|
-Supramolecule #1: mTORC2 in complex with its regulator DEPTOR
| Supramolecule | Name: mTORC2 in complex with its regulator DEPTOR / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 1.24 MDa |
-Macromolecule #1: Serine/threonine-protein kinase mTOR
| Macromolecule | Name: Serine/threonine-protein kinase mTOR / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 291.321938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLGTGPAAAT TAATTSSNVS VLQQFASGLK SRNEETRAKA AKELQHYVTM ELREMSQEES TRFYDQLNHH IFELVSSSDA NERKGGILA IASLIGVEGG NATRIGRFAN YLRNLLPSND PVVMEMASKA IGRLAMAGDT FTAEYVEFEV KRALEWLGAD R NEGRRHAA ...String: MLGTGPAAAT TAATTSSNVS VLQQFASGLK SRNEETRAKA AKELQHYVTM ELREMSQEES TRFYDQLNHH IFELVSSSDA NERKGGILA IASLIGVEGG NATRIGRFAN YLRNLLPSND PVVMEMASKA IGRLAMAGDT FTAEYVEFEV KRALEWLGAD R NEGRRHAA VLVLRELAIS VPTFFFQQVQ PFFDNIFVAV WDPKQAIREG AVAALRACLI LTTQREPKEM QKPQWYRHTF EE AEKGFDE TLAKEKGMNR DGSTSGSGDY KDDDDKGSTS GSGDRIHGAL LILNELVRIS SMEGERLREE MEEITQQQLV HDK YCKDLM GFGTKPRHIT PFTSFQAVQP QQSNALVGLL GYSSHQGLMG FGTSPSPAKS TLVESRCCRD LMEEKFDQVC QWVL KCRNS KNSLIQMTIL NLLPRLAAFR PSAFTDTQYL QDTMNHVLSC VKKEKERTAA FQALGLLSVA VRSEFKVYLP RVLDI IRAA LPPKDFAHKR QKAMQVDATV FTCISMLARA MGPGIQQDIK ELLEPMLAVG LSPALTAVLY DLSRQIPQLK KDIQDG LLK MLSLVLMHKP LRHPGMPKGL AHQLASPGLT TLPEASDVGS ITLALRTLGS FEFEGHSLTQ FVRHCADHFL NSEHKEI RM EAARTCSRLL TPSIHLISGH AHVVSQTAVQ VVADVLSKLL VVGITDPDPD IRYCVLASLD ERFDAHLAQA ENLQALFV A LNDQVFEIRE LAICTVGRLS SMNPAFVMPF LRKMLIQILT ELEHSGIGRI KEQSARMLGH LVSNAPRLIR PYMEPILKA LILKLKDPDP DPNPGVINNV LATIGELAQV SGLEMRKWVD ELFIIIMDML QDSSLLAKRQ VALWTLGQLV ASTGYVVEPY RKYPTLLEV LLNFLKTEQN QGTRREAIRV LGLLGALDPY KHKVNIGMID QSRDASAVSL SESKSSQDSS DYSTSEMLVN M GNLPLDEF YPAVSMVALM RIFRDQSLSH HHTMVVQAIT FIFKSLGLKC VQFLPQVMPT FLNVIRVCDG AIREFLFQQL GM LVSFVKS HIRPYMDEIV TLMREFWVMN TSIQSTIILL IEQIVVALGG EFKLYLPQLI PHMLRVFMHD NSPGRIVSIK LLA AIQLFG ANLDDYLHLL LPPIVKLFDA PEAPLPSRKA ALETVDRLTE SLDFTDYASR IIHPIVRTLD QSPELRSTAM DTLS SLVFQ LGKKYQIFIP MVNKVLVRHR INHQRYDVLI CRIVKGYTLA DEEEDPLIYQ HRMLRSGQGD ALASGPVETG PMKKL HVST INLQKAWGAA RRVSKDDWLE WLRRLSLELL KDSSSPSLRS CWALAQAYNP MARDLFNAAF VSCWSELNED QQDELI RSI ELALTSQDIA EVTQTLLNLA EFMEHSDKGP LPLRDDNGIV LLGERAAKCR AYAKALHYKE LEFQKGPTPA ILESLIS IN NKLQQPEAAA GVLEYAMKHF GELEIQATWY EKLHEWEDAL VAYDKKMDTN KDDPELMLGR MRCLEALGEW GQLHQQCC E KWTLVNDETQ AKMARMAAAA AWGLGQWDSM EEYTCMIPRD THDGAFYRAV LALHQDLFSL AQQCIDKARD LLDAELTAM AGESYSRAYG AMVSCHMLSE LEEVIQYKLV PERREIIRQI WWERLQGCQR IVEDWQKILM VRSLVVSPHE DMRTWLKYAS LCGKSGRLA LAHKTLVLLL GVDPSRQLDH PLPTVHPQVT YAYMKNMWKS ARKIDAFQHM QHFVQTMQQQ AQHAIATEDQ Q HKQELHKL MARCFLKLGE WQLNLQGINE STIPKVLQYY SAATEHDRSW YKAWHAWAVM NFEAVLHYKH QNQARDEKKK LR HASGANI TNATTAATTA ATATTTASTE GSNSESEAES TENSPTPSPL QKKVTEDLSK TLLMYTVPAV QGFFRSISLS RGN NLQDTL RVLTLWFDYG HWPDVNEALV EGVKAIQIDT WLQVIPQLIA RIDTPRPLVG RLIHQLLTDI GRYHPQALIY PLTV ASKST TTARHNAANK ILKNMCEHSN TLVQQAMMVS EELIRVAILW HEMWHEGLEE ASRLYFGERN VKGMFEVLEP LHAMM ERGP QTLKETSFNQ AYGRDLMEAQ EWCRKYMKSG NVKDLTQAWD LYYHVFRRIS KQLPQLTSLE LQYVSPKLLM CRDLEL AVP GTYDPNQPII RIQSIAPSLQ VITSKQRPRK LTLMGSNGHE FVFLLKGHED LRQDERVMQL FGLVNTLLAN DPTSLRK NL SIQRYAVIPL STNSGLIGWV PHCDTLHALI RDYREKKKIL LNIEHRIMLR MAPDYDHLTL MQKVEVFEHA VNNTAGDD L AKLLWLKSPS SEVWFDRRTN YTRSLAVMSM VGYILGLGDR HPSNLMLDRL SGKILHIDFG DCFEVAMTRE KFPEKIPFR LTRMLTNAME VTGLDGNYRI TCHTVMEVLR EHKDSVMAVL EAFVYDPLLN WRLMDTNTKG NKRSRTRTDS YSAGQSVEIL DGVELGEPA HKKTGTTVPE SIHSFIGDGL VKPEALNKKA IQIINRVRDK LTGRDFSHDD TLDVPTQVEL LIKQATSHEN L CQCYIGWC PFW |
-Macromolecule #2: Target of rapamycin complex subunit LST8
| Macromolecule | Name: Target of rapamycin complex subunit LST8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.91009 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNTSPGTVGS DPVILATAGY DHTVRFWQAH SGICTRTVQH QDSQVNALEV TPDRSMIAAA GYQHIRMYDL NSNNPNPIIS YDGVNKNIA SVGFHEDGRW MYTGGEDCTA RIWDLRSRNL QCQRIFQVNA PINCVCLHPN QAELIVGDQS GAIHIWDLKT D HNEQLIPE ...String: MNTSPGTVGS DPVILATAGY DHTVRFWQAH SGICTRTVQH QDSQVNALEV TPDRSMIAAA GYQHIRMYDL NSNNPNPIIS YDGVNKNIA SVGFHEDGRW MYTGGEDCTA RIWDLRSRNL QCQRIFQVNA PINCVCLHPN QAELIVGDQS GAIHIWDLKT D HNEQLIPE PEVSITSAHI DPDASYMAAV NSTGNCYVWN LTGGIGDEVT QLIPKTKIPA HTRYALQCRF SPDSTLLATC SA DQTCKIW RTSNFSLMTE LSIKSGNPGE SSRGWMWGCA FSGDSQYIVT ASSDNLARLW CVETGEIKRE YGGHQKAVVC LAF NDSVLG |
-Macromolecule #3: Rapamycin-insensitive companion of mTOR
| Macromolecule | Name: Rapamycin-insensitive companion of mTOR / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 192.472922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAIGRGRSL KNLRVRGRND SGEENVPLDL TREPSDNLRE ILQNVARLQG VSNMRKLGHL NNFTKLLCDI GHSEEKLGFH YEDIIICLR LALLNEAKEV RAAGLRALRY LIQDSSILQK VLKLKVDYLI ARCIDIQQSN EVERTQALRL VRKMITVNAS L FPSSVTNS ...String: MAAIGRGRSL KNLRVRGRND SGEENVPLDL TREPSDNLRE ILQNVARLQG VSNMRKLGHL NNFTKLLCDI GHSEEKLGFH YEDIIICLR LALLNEAKEV RAAGLRALRY LIQDSSILQK VLKLKVDYLI ARCIDIQQSN EVERTQALRL VRKMITVNAS L FPSSVTNS LIAVGNDGLQ ERDRMVRACI AIICELALQN PEVVALRGGL NTILKNVIDC QLSRINEALI TTILHLLNHP KT RQYVRAD VELERILAPY TDFHYRHSPD TAEGQLKEDR EARFLASKMG IIATFRSWAG IINLCKPGNS GIQSLIGVLC IPN MEIRRG LLEVLYDIFR LPLPVVTEEF IEALLSVDPG RFQDSWRLSD GFVAAEAKTI LPHRARSRPD LMDNYLALIL SAFI RNGLL EGLVEVITNS DDHISVRATI LLGELLHMAN TILPHSHSHH LHCLPTLMNM AASFDIPKEK RLRASAALNC LKRFH EMKK RGPKPYSLHL DHIIQKAIAT HQKRDQYLRV QKDIFILKDT EEALLINLRD SQVLQHKENL EWNWNLIGTI LKWPNV NLR NYKDEQLHRF VRRLLYFYKP SSKLYANLDL DFAKAKQLTV VGCQFTEFLL ESEEDGQGYL EDLVKDIVQW LNASSGM KP ERSLQNNGLL TTLSQHYFLF IGTLSCHPHG VKMLEKCSVF QCLLNLCSLK NQDHLLKLTV SSLDYSRDGL ARVILSKI L TAATDACRLY ATKHLRVLLR ANVEFFNNWG IELLVTQLHD KNKTISSEAL DILDEACEDK ANLHALIQMK PALSHLGDK GLLLLLRFLS IPKGFSYLNE RGYVAKQLEK WHREYNSKYV DLIEEQLNEA LTTYRKPVDG DNYVRRSNQR LQRPHVYLPI HLYGQLVHH KTGCHLLEVQ NIITELCRNV RTPDLDKWEE IKKLKASLWA LGNIGSSNWG LNLLQEENVI PDILKLAKQC E VLSIRGTC VYVLGLIAKT KQGCDILKCH NWDAVRHSRK HLWPVVPDDV EQLCNELSSI PSTLSLNSES TSSRHNSESE SV PSSMFIL EDDRFGSSST STFFLDINED TEPTFYDRSG PIKDKNSFPF FASSKLVKNR ILNSLTLPNK KHRSSSDPKG GKL SSESKT SNRRIRTLTE PSVDFNHSDD FTPISTVQKT LQLETSFMGN KHIEDTGSTP SIGENDLKFT KNFGTENHRE NTSR ERLVV ESSTSSHMKI RSQSFNTDTT TSGISSMSSS PSRETVGVDA TTMDTDCGSM STVVSTKTIK TSHYLTPQSN HLSLS KSNS VSLVPPGSSH TLPRRAQSLK APSIATIKSL ADCNFSYTSS RDAFGYATLK RLQQQRMHPS LSHSEALASP AKDVLF TDT ITMKANSFES RLTPSRFMKA LSYASLDKED LLSPINQNTL QRSSSVRSMV SSATYGGSDD YIGLALPVDI NDIFQVK DI PYFQTKNIPP HDDRGARAFA HDAGGLPSGT GGLVKNSFHL LRQQMSLTEI MNSIHSDASL FLESTEDTGL QEHTDDNC L YCVCIEILGF QPSNQLSAIC SHSDFQDIPY SDWCEQTIHN PLEVVPSKFS GISGCSDGVS QEGSASSTKS TELLLGVKT IPDDTPMCRI LLRKEVLRLV INLSSSVSTK CHETGLLTIK EKYPQTFDDI CLYSEVSHLL SHCTFRLPCR RFIQELFQDV QFLQMHEEA EAVLATPPKQ PIVDTSAES |
-Macromolecule #4: Target of rapamycin complex 2 subunit MAPKAP1
| Macromolecule | Name: Target of rapamycin complex 2 subunit MAPKAP1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.206738 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAFLDNPTII LAHIRQSHVT SDDTGMCEMV LIDHDVDLEK IHPPSMPGDS GSEIQGSNGE TQGYVYAQSV DITSSWDFGI RRRSNTAQR LERLRKERQN QIKCKNIQWK ERNSKQSAQE LKSLFEKKSL KEKPPISGKQ SILSVRLEQC PLQLNNPFNE Y SKFDGKGH ...String: MAFLDNPTII LAHIRQSHVT SDDTGMCEMV LIDHDVDLEK IHPPSMPGDS GSEIQGSNGE TQGYVYAQSV DITSSWDFGI RRRSNTAQR LERLRKERQN QIKCKNIQWK ERNSKQSAQE LKSLFEKKSL KEKPPISGKQ SILSVRLEQC PLQLNNPFNE Y SKFDGKGH VGTTATKKID VYLPLHSSQD RLLPMTVVTM ASARVQDLIG LICWQYTSEG REPKLNDNVS AYCLHIAEDD GE VDTDFPP LDSNEPIHKF GFSTLALVEK YSSPGLTSKE SLFVRINAAH GFSLIQVDNT KVTMKEILLK AVKRRKGSQK VSG PQYRLE KQSEPNVAVD LDSTLESQSA WEFCLVRENS SRADGVFEED SQIDIATVQD MLSSHHYKSF KVSMIHRLRF TTDV QLGIS GDKVEIDPVT NQKASTKFWI KQKPISIDSD LLCACDLAEE KSPSHAIFKL TYLSNHDYKH LYFESDAATV NEIVL KVNY ILESRASTAR ADYFAQKQRK LNRRTSFSFQ KEKKSGQQ |
-Macromolecule #5: DEP domain-containing mTOR-interacting protein
| Macromolecule | Name: DEP domain-containing mTOR-interacting protein / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.365832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEEGGSTGSA GSDSSTSGSG GAQQRELERM AEVLVTGEQL RLRLHEEKVI KDRRHHLKTY PNCFVAKELI DWLIEHKEAS DRETAIKLM QKLADRGIIH HVCDEHKEFK DVKLFYRFRK DDGTFPLDNE VKAFMRGQRL YEKLMSPENT LLQPREEEGV K YERTFMAS ...String: MEEGGSTGSA GSDSSTSGSG GAQQRELERM AEVLVTGEQL RLRLHEEKVI KDRRHHLKTY PNCFVAKELI DWLIEHKEAS DRETAIKLM QKLADRGIIH HVCDEHKEFK DVKLFYRFRK DDGTFPLDNE VKAFMRGQRL YEKLMSPENT LLQPREEEGV K YERTFMAS EFLDWLVQEG EATTRKEAEQ LCHRLMEHGI IQHVSSKHPF VDSNLLYQFR MNFRRRRRLM ELLNEKSPSS QE THDSPFC LRKQSHDNRK STSFMSVSPS KEIKIVSAVR RSSMSSCGSS GYFSSSPTLS SSPPVLCNPK SVLKRPVTSE ELL TPGAPY ARKTFTIVGD AVGWGFVVRG SKPCHIQAVD PSGPAAAAGM KVCQFVVSVN GLNVLHVDYR TVNNLILTGP RTIV MEVME ELEC |
-Macromolecule #6: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #8: ACETYL GROUP
| Macromolecule | Name: ACETYL GROUP / type: ligand / ID: 8 / Number of copies: 1 / Formula: ACE |
|---|---|
| Molecular weight | Theoretical: 44.053 Da |
| Chemical component information |  ChemComp-ACE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: RELION (ver. 3.1) |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.2.0) / Number images used: 750254 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT / Software - Name: cryoSPARC (ver. 3.2.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.2.0) |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)