+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of PriRep1 with dsDNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helicase / SaPI1 / REPLICATION | |||||||||
| Function / homology | Virulence-associated E / Virulence-associated protein E-like domain / P-loop containing nucleoside triphosphate hydrolase / Similar to D. nodosus vapE Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
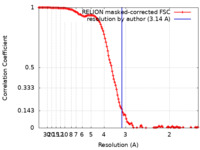
| Method | single particle reconstruction / cryo EM / Resolution: 3.14 Å | |||||||||
 Authors Authors | Qiao CC / Mir Sanchis I | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility. Authors: Cuncun Qiao / Gianluca Debiasi-Anders / Ignacio Mir-Sanchis /  Abstract: Replication is a crucial cellular process. Replicative helicases unwind DNA providing the template strand to the polymerase and promoting replication fork progression. Helicases are multi-domain ...Replication is a crucial cellular process. Replicative helicases unwind DNA providing the template strand to the polymerase and promoting replication fork progression. Helicases are multi-domain proteins which use an ATPase domain to couple ATP hydrolysis with translocation, however the role that the other domains might have during translocation remains elusive. Here, we studied the unexplored self-loading helicases called Reps, present in Staphylococcus aureus pathogenicity islands (SaPIs). Our cryoEM structures of the PriRep5 from SaPI5 (3.3 Å), the Rep1 from SaPI1 (3.9 Å) and Rep1-DNA complex (3.1Å) showed that in both Reps, the C-terminal domain (CTD) undergoes two distinct movements respect the ATPase domain. We experimentally demonstrate both in vitro and in vivo that SaPI-encoded Reps need key amino acids involved in the staircase mechanism of translocation. Additionally, we demonstrate that the CTD's presence is necessary for the maintenance of full ATPase and helicase activities. We speculate that this high interdomain flexibility couples Rep's activities as initiators and as helicases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13342.map.gz emd_13342.map.gz | 166.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13342-v30.xml emd-13342-v30.xml emd-13342.xml emd-13342.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13342_fsc.xml emd_13342_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13342.png emd_13342.png | 136.7 KB | ||
| Masks |  emd_13342_msk_1.map emd_13342_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13342.cif.gz emd-13342.cif.gz | 6.2 KB | ||
| Others |  emd_13342_half_map_1.map.gz emd_13342_half_map_1.map.gz emd_13342_half_map_2.map.gz emd_13342_half_map_2.map.gz | 140.7 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13342 http://ftp.pdbj.org/pub/emdb/structures/EMD-13342 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13342 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13342 | HTTPS FTP |
-Related structure data
| Related structure data |  7pdsMC  7olaC  7om0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-10880 (Title: Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility EMPIAR-10880 (Title: Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibilityData size: 395.0 / Data #1: PriRep1-DNA [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13342.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13342.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13342_msk_1.map emd_13342_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13342_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13342_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PriRep1-dsDNA
| Entire | Name: PriRep1-dsDNA |
|---|---|
| Components |
|
-Supramolecule #1: PriRep1-dsDNA
| Supramolecule | Name: PriRep1-dsDNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Similar to D. nodosus vapE
| Macromolecule | Name: Similar to D. nodosus vapE / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.423895 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFEMIDSRTG VLNANDWKSQ LRRSATTQAL KKTTTNAEII LCNDESLKGL VQYDAFEKVT KLKRLPYWRS KGDANYYWAD IDTTHVISH IDKLYNVQFS RDLIDTVIEK EAYQNRFHPI KSMIESKSWD GIKRIETLFI DYLGAEDNHY NREVTKKWMM G AVARIYQP ...String: MFEMIDSRTG VLNANDWKSQ LRRSATTQAL KKTTTNAEII LCNDESLKGL VQYDAFEKVT KLKRLPYWRS KGDANYYWAD IDTTHVISH IDKLYNVQFS RDLIDTVIEK EAYQNRFHPI KSMIESKSWD GIKRIETLFI DYLGAEDNHY NREVTKKWMM G AVARIYQP GIKYDSMIIL YGGQGVGKST AVSKLGGHWY NQSIKTFKGD EVYKKLQGSW ICEIEELSAF QKSTIEDIKG FI SAIVDIY RASYGKRTER HPRQCVFVGT TNNYEFLKDQ TGNRRFFPIT TDKNKATKSP FDDLTPVVVQ QMFAEARVYF DEN PTDKAL LLDKEASEMA LKVQEAHSEK DALVGEIEEF LERPIPSDYW YRTLEEKRVS AHDVIDQDYI KLYGDGKLIE LPNA KPGAY VWRDKVCSME IWKVMMKRDD QPQQHHLRKI DKALRNTNYC GTVKKQTRYG EGIGKQYGFS VDLASYYKNL KV UniProtKB: Similar to D. nodosus vapE |
-Macromolecule #2: polyA
| Macromolecule | Name: polyA / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.512063 KDa |
| Sequence | String: (DT)(DA)(DA)(DA)(DA) |
-Macromolecule #3: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 6 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.22 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 20.0 mM/L / Component - Formula: Tris |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 16 / Number real images: 3121 / Average exposure time: 5.0 sec. / Average electron dose: 1.44 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: correction coefficient |
|---|---|
| Output model |  PDB-7pds: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)