[English] 日本語
 Yorodumi
Yorodumi- EMDB-1318: Three-dimensional structure of the respiratory chain supercomplex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1318 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. | |||||||||
 Map data Map data | This is a side view along the mitochondiral membrane plane | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 32.0 Å | |||||||||
 Authors Authors | Schafer E / Dencher NA / Vonck J / Parcej DN | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2007 Journal: Biochemistry / Year: 2007Title: Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Authors: Eva Schäfer / Norbert A Dencher / Janet Vonck / David N Parcej /  Abstract: The respiratory chain complexes can arrange into multienzyme assemblies, so-called supercomplexes. We present the first 3D map of a respiratory chain supercomplex. It was determined by random conical ...The respiratory chain complexes can arrange into multienzyme assemblies, so-called supercomplexes. We present the first 3D map of a respiratory chain supercomplex. It was determined by random conical tilt electron microscopy analysis of a bovine supercomplex consisting of complex I, dimeric complex III, and complex IV (I1III2IV1). Within this 3D map the positions and orientations of all the individual complexes in the supercomplex were determined unambiguously. Furthermore, the ubiquinone and cytochrome c binding sites of each complex in the supercomplex could be located. The mobile electron carrier binding site of each complex was found to be in proximity to the binding site of the succeeding complex in the respiratory chain. This provides structural evidence for direct substrate channeling in the supercomplex assembly with short diffusion distances for the mobile electron carriers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1318.map.gz emd_1318.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1318-v30.xml emd-1318-v30.xml emd-1318.xml emd-1318.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  1318.gif 1318.gif | 35.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1318 http://ftp.pdbj.org/pub/emdb/structures/EMD-1318 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1318 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1318 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1318.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1318.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a side view along the mitochondiral membrane plane | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.667 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
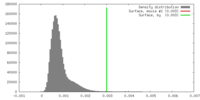
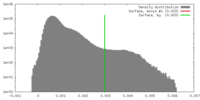
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : bovine supercomplex I1III2IV1
| Entire | Name: bovine supercomplex I1III2IV1 |
|---|---|
| Components |
|
-Supramolecule #1000: bovine supercomplex I1III2IV1
| Supramolecule | Name: bovine supercomplex I1III2IV1 / type: sample / ID: 1000 Details: membranes are solubilised in digitonin, monodisperse sample Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 1.7 MDa / Theoretical: 1.7 MDa |
-Macromolecule #1: complex I
| Macromolecule | Name: complex I / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #2: complex III
| Macromolecule | Name: complex III / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Oligomeric state: dimer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #3: complex IV
| Macromolecule | Name: complex IV / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.1 Details: 0.1 % (w/v) digitonin, 25 mM tricine, 7.5 mM bis-tris, 25 mM aminocaproic acid, 10 % (w/v) glycerol |
| Staining | Type: NEGATIVE / Details: 2 % (w/v) ammonium molybdate, deep stain method |
| Grid | Details: 400 mesh Cu grid |
| Vitrification | Cryogen name: NONE |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM120T |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Number real images: 20 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder: n.a / Specimen holder model: OTHER / Tilt angle max: 50 |
- Image processing
Image processing
| Details | particles were selected manually |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 32.0 Å / Resolution method: FSC 3 SIGMA CUT-OFF / Software - Name: SPIDER / Details: final maps calculated from two / Number images used: 1023 |
| Final two d classification | Number classes: 3 |
-Atomic model buiding 1
| Details | The complexes were fitted by hand in Chimera |
|---|
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)