[English] 日本語
 Yorodumi
Yorodumi- EMDB-13179: Streptomyces coelicolor dATP/ATP-loaded NrdR in complex with its ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Streptomyces coelicolor dATP/ATP-loaded NrdR in complex with its cognate DNA | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Repressor / Dodecamer / ATP-binding / dATP-binding / DNA BINDING PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationdouble-stranded DNA binding / negative regulation of DNA-templated transcription / zinc ion binding / ATP binding Similarity search - Function | ||||||||||||||||||
| Biological species |  Streptomyces coelicolor A3(2) (bacteria) / Streptomyces coelicolor A3(2) (bacteria) /  Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) (bacteria) Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) (bacteria) | ||||||||||||||||||
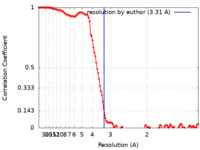
| Method | single particle reconstruction / cryo EM / Resolution: 3.31 Å | ||||||||||||||||||
 Authors Authors | Martinez-Carranza M / Stenmark P | ||||||||||||||||||
| Funding support |  Sweden, 5 items Sweden, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases. Authors: Inna Rozman Grinberg / Markel Martínez-Carranza / Ornella Bimai / Ghada Nouaïria / Saher Shahid / Daniel Lundin / Derek T Logan / Britt-Marie Sjöberg / Pål Stenmark /  Abstract: Ribonucleotide reductase (RNR) is an essential enzyme that catalyzes the synthesis of DNA building blocks in virtually all living cells. NrdR, an RNR-specific repressor, controls the transcription of ...Ribonucleotide reductase (RNR) is an essential enzyme that catalyzes the synthesis of DNA building blocks in virtually all living cells. NrdR, an RNR-specific repressor, controls the transcription of RNR genes and, often, its own, in most bacteria and some archaea. NrdR senses the concentration of nucleotides through its ATP-cone, an evolutionarily mobile domain that also regulates the enzymatic activity of many RNRs, while a Zn-ribbon domain mediates binding to NrdR boxes upstream of and overlapping the transcription start site of RNR genes. Here, we combine biochemical and cryo-EM studies of NrdR from Streptomyces coelicolor to show, at atomic resolution, how NrdR binds to DNA. The suggested mechanism involves an initial dodecamer loaded with two ATP molecules that cannot bind to DNA. When dATP concentrations increase, an octamer forms that is loaded with one molecule each of dATP and ATP per monomer. A tetramer derived from this octamer then binds to DNA and represses transcription of RNR. In many bacteria - including well-known pathogens such as Mycobacterium tuberculosis - NrdR simultaneously controls multiple RNRs and hence DNA synthesis, making it an excellent target for novel antibiotics development. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13179.map.gz emd_13179.map.gz | 153.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13179-v30.xml emd-13179-v30.xml emd-13179.xml emd-13179.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13179_fsc.xml emd_13179_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13179.png emd_13179.png | 63.9 KB | ||
| Filedesc metadata |  emd-13179.cif.gz emd-13179.cif.gz | 6.3 KB | ||
| Others |  emd_13179_half_map_1.map.gz emd_13179_half_map_1.map.gz emd_13179_half_map_2.map.gz emd_13179_half_map_2.map.gz | 151.3 MB 151.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13179 http://ftp.pdbj.org/pub/emdb/structures/EMD-13179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13179 | HTTPS FTP |
-Related structure data
| Related structure data |  7p3fMC  7p37C  7p3qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13179.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13179.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.67 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_13179_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
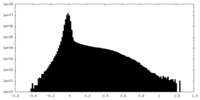
| Projections & Slices |
| ||||||||||||
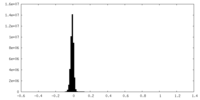
| Density Histograms |
-Half map: #1
| File | emd_13179_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
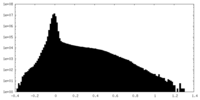
| Projections & Slices |
| ||||||||||||
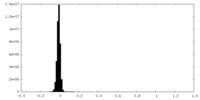
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric assembly of dATP/ATP-loaded NrdR in complex with its c...
| Entire | Name: Tetrameric assembly of dATP/ATP-loaded NrdR in complex with its cognate DNA |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric assembly of dATP/ATP-loaded NrdR in complex with its c...
| Supramolecule | Name: Tetrameric assembly of dATP/ATP-loaded NrdR in complex with its cognate DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 125 KDa |
-Supramolecule #2: Transcriptional repressor NrdR
| Supramolecule | Name: Transcriptional repressor NrdR / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
-Supramolecule #3: DNA (50-MER)
| Supramolecule | Name: DNA (50-MER) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
-Macromolecule #1: Transcriptional repressor NrdR
| Macromolecule | Name: Transcriptional repressor NrdR / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) (bacteria) Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) (bacteria)Strain: ATCC BAA-471 / A3(2) / M145 |
| Molecular weight | Theoretical: 21.271629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHCPFCRHPD SRVVDSRTTD DGTSIRRRRQ CPDCSRRFTT VETCSLMVVK RSGVTEPFSR TKVINGVRKA CQGRPVTEDA LAQLGQRVE EAVRATGSAE LTTHDVGLAI LGPLQELDLV AYLRFASVYR AFDSLEDFEA AIAELRETTG HPGEEDDTGA G SQENDRGP ...String: MHCPFCRHPD SRVVDSRTTD DGTSIRRRRQ CPDCSRRFTT VETCSLMVVK RSGVTEPFSR TKVINGVRKA CQGRPVTEDA LAQLGQRVE EAVRATGSAE LTTHDVGLAI LGPLQELDLV AYLRFASVYR AFDSLEDFEA AIAELRETTG HPGEEDDTGA G SQENDRGP TGAGQVPEPA GAADKLAAAL EHHHHHH UniProtKB: Transcriptional repressor NrdR |
-Macromolecule #2: DNA (50-MER)
| Macromolecule | Name: DNA (50-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Molecular weight | Theoretical: 17.507195 KDa |
| Sequence | String: (DG)(DC)(DC)(DA)(DA)(DT)(DC)(DC)(DC)(DC) (DA)(DC)(DA)(DT)(DC)(DT)(DA)(DG)(DT)(DG) (DG)(DT)(DT)(DG)(DG)(DA)(DT)(DA)(DG) (DC)(DG)(DT)(DG)(DA)(DG)(DC)(DA)(DG)(DC) (DC) (DC)(DA)(DC)(DA)(DA)(DG) ...String: (DG)(DC)(DC)(DA)(DA)(DT)(DC)(DC)(DC)(DC) (DA)(DC)(DA)(DT)(DC)(DT)(DA)(DG)(DT)(DG) (DG)(DT)(DT)(DG)(DG)(DA)(DT)(DA)(DG) (DC)(DG)(DT)(DG)(DA)(DG)(DC)(DA)(DG)(DC) (DC) (DC)(DA)(DC)(DA)(DA)(DG)(DT)(DT) (DG)(DT)(DG)(DG)(DT)(DC)(DC)(DC)(DC) |
-Macromolecule #3: DNA (50-MER)
| Macromolecule | Name: DNA (50-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Molecular weight | Theoretical: 17.627266 KDa |
| Sequence | String: (DG)(DG)(DG)(DG)(DA)(DC)(DC)(DA)(DC)(DA) (DA)(DC)(DT)(DT)(DG)(DT)(DG)(DG)(DG)(DC) (DT)(DG)(DC)(DT)(DC)(DA)(DC)(DG)(DC) (DT)(DA)(DT)(DC)(DC)(DA)(DA)(DC)(DC)(DA) (DC) (DT)(DA)(DG)(DA)(DT)(DG) ...String: (DG)(DG)(DG)(DG)(DA)(DC)(DC)(DA)(DC)(DA) (DA)(DC)(DT)(DT)(DG)(DT)(DG)(DG)(DG)(DC) (DT)(DG)(DC)(DT)(DC)(DA)(DC)(DG)(DC) (DT)(DA)(DT)(DC)(DC)(DA)(DA)(DC)(DC)(DA) (DC) (DT)(DA)(DG)(DA)(DT)(DG)(DT)(DG) (DG)(DG)(DG)(DA)(DT)(DT)(DG)(DG)(DC) |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 4 / Formula: DTP |
|---|---|
| Molecular weight | Theoretical: 491.182 Da |
| Chemical component information |  ChemComp-DTP: |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3562 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)